Abstract
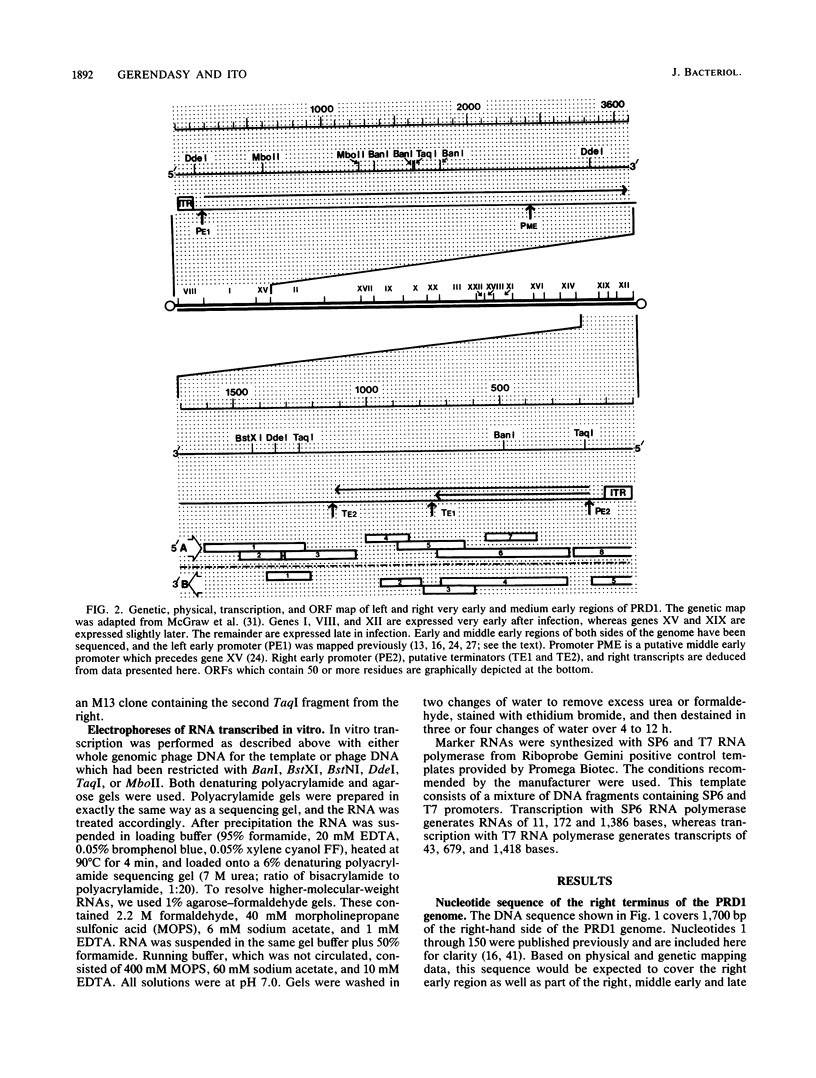
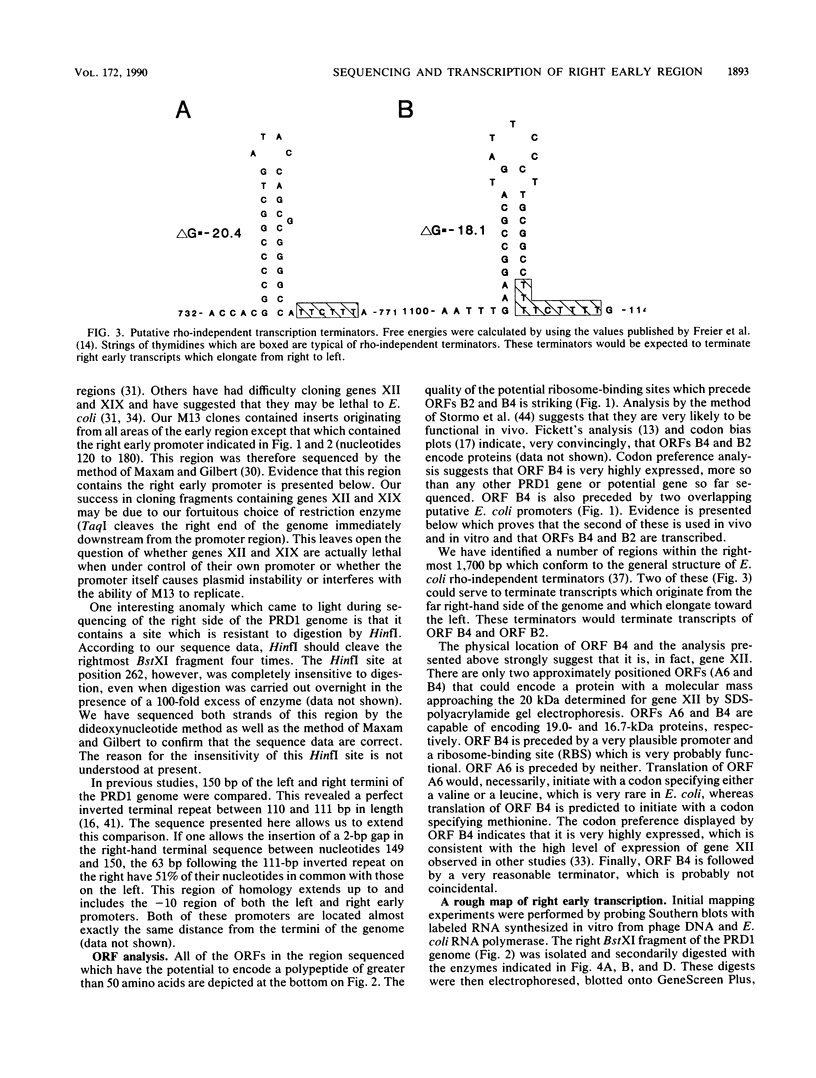
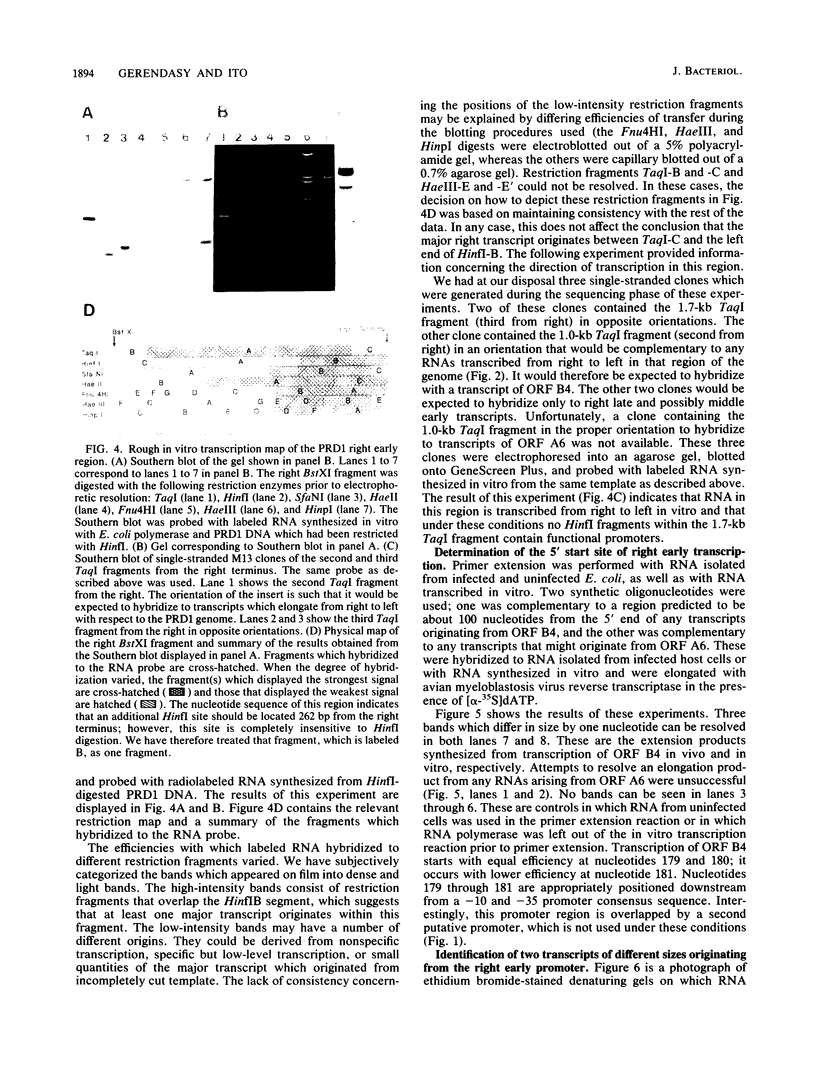
We have sequenced the rightmost 1,700 base pairs of bacteriophage PRD1. This region encompasses the right early region and completes the sequence of all PRD1 early functions. We have also mapped the 5' initiation site of right early transcripts in vivo and in vitro. This has allowed us to assign gene XII to an open reading frame and suggests that another open reading frame may also be expressed. Gene XII, which has been implicated in the replication process and the regulation of gene expression, is predicted to encode a protein with a molecular mass of 16.7 kilodaltons. Data base searches have revealed no significant homology between the product of this gene and other proteins. Transcription mapping studies have revealed that right early transcripts elongate from right to left and have enabled us to identify the right early promoter. This promoter behaves identically in vivo and in vitro. We also demonstrate that this promoter directs the transcription of two RNAs of different sizes in vitro.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Adhya S., de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli cells. J Biol Chem. 1981 Nov 25;256(22):11905–11910. [PubMed] [Google Scholar]
- Bamford D. H., Mindich L. Characterization of the DNA-protein complex at the termini of the bacteriophage PRD1 genome. J Virol. 1984 May;50(2):309–315. doi: 10.1128/jvi.50.2.309-315.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D. E., Rutherford E. L. Basic characterization of a lipid-containing bacteriophage specific for plasmids of the P, N, and W compatibility groups. Can J Microbiol. 1975 Feb;21(2):152–163. doi: 10.1139/m75-023. [DOI] [PubMed] [Google Scholar]
- Chase J. W., Williams K. R. Single-stranded DNA binding proteins required for DNA replication. Annu Rev Biochem. 1986;55:103–136. doi: 10.1146/annurev.bi.55.070186.000535. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Qasba P. K. Alkaline transfer of DNA to plastic membrane. Biochem Biophys Res Commun. 1984 Jul 18;122(1):340–344. doi: 10.1016/0006-291x(84)90480-7. [DOI] [PubMed] [Google Scholar]
- Davis T. N., Muller E. D., Cronan J. E., Jr The virion of the lipid-containing bacteriophage PR4. Virology. 1982 Jul 30;120(2):287–306. doi: 10.1016/0042-6822(82)90031-9. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Weiss R. M., Terwilliger T. C. The helical hydrophobic moment: a measure of the amphiphilicity of a helix. Nature. 1982 Sep 23;299(5881):371–374. doi: 10.1038/299371a0. [DOI] [PubMed] [Google Scholar]
- Escarmis C., García P., Méndez E., López R., Salas M., García E. Inverted terminal repeats and terminal proteins of the genomes of pneumococcal phages. Gene. 1985;36(3):341–348. doi: 10.1016/0378-1119(85)90189-1. [DOI] [PubMed] [Google Scholar]
- Escarmís C., Gómez A., García E., Ronda C., López R., Salas M. Nucleotide sequence at the termini of the DNA of Streptococcus pneumoniae phage Cp-1. Virology. 1984 Feb;133(1):166–171. doi: 10.1016/0042-6822(84)90435-5. [DOI] [PubMed] [Google Scholar]
- Fickett J. W. Recognition of protein coding regions in DNA sequences. Nucleic Acids Res. 1982 Sep 11;10(17):5303–5318. doi: 10.1093/nar/10.17.5303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentz R., Bujard H. Promoters recognized by Escherichia coli RNA polymerase selected by function: highly efficient promoters from bacteriophage T5. J Bacteriol. 1985 Oct;164(1):70–77. doi: 10.1128/jb.164.1.70-77.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerendasy D. D., Ito J. The genome of lipid-containing bacteriophage PRD1, which infects gram-negative bacteria, contains long, inverted terminal repeats. J Virol. 1987 Feb;61(2):594–596. doi: 10.1128/jvi.61.2.594-596.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Devereux J., Burgess R. R. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. X., Bailey S., Bodley J. W., Anderson D. Characterization of the small RNA of the bacteriophage phi 29 DNA packaging machine. Nucleic Acids Res. 1987 Sep 11;15(17):7081–7090. doi: 10.1093/nar/15.17.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P. X., Erickson S., Anderson D. A small viral RNA is required for in vitro packaging of bacteriophage phi 29 DNA. Science. 1987 May 8;236(4802):690–694. doi: 10.1126/science.3107124. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. C., Jung G. H., Leavitt M. C., Ito J. Primary structure of the DNA terminal protein of bacteriophage PRD1. Nucleic Acids Res. 1987 Nov 11;15(21):8999–9009. doi: 10.1093/nar/15.21.8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys A. J., Flavell R. A. A physical map of the DNA regions flanking the rabbit beta-globin gene. Cell. 1977 Oct;12(2):429–439. doi: 10.1016/0092-8674(77)90119-2. [DOI] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Hsieh J. C., Ito J. Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8287–8291. doi: 10.1073/pnas.84.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura F., Ito J. Transcription of the genome of bacteriophage phi 29: isolation and mapping of the major early mRNA synthesized in vivo and in vitro. J Virol. 1977 Sep;23(3):562–577. doi: 10.1128/jvi.23.3.562-577.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Lundström K. H., Bamford D. H., Palva E. T., Lounatmaa K. Lipid-containing bacteriophage PR4: structure and life cycle. J Gen Virol. 1979 Jun;43(3):583–592. doi: 10.1099/0022-1317-43-3-583. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McGraw T., Yang H. L., Mindich L. Establishment of a physical and genetic map for bacteriophage PRD1. Mol Gen Genet. 1983;190(2):237–244. doi: 10.1007/BF00330646. [DOI] [PubMed] [Google Scholar]
- Mellado R. P., Moreno F., Viñuela E., Salas M., Reilly B. E., Anderson D. L. Genetic analysis of bacteriophage phi 29 of Bacillus subtilis: integration and mapping of reference mutants of two collections. J Virol. 1976 Aug;19(2):495–500. doi: 10.1128/jvi.19.2.495-500.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindich L., Bamford D., Goldthwaite C., Laverty M., Mackenzie G. Isolation of nonsense mutants of lipid-containing bacteriophage PRD1. J Virol. 1982 Dec;44(3):1013–1020. doi: 10.1128/jvi.44.3.1013-1020.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Olsen R. H., Siak J. S., Gray R. H. Characteristics of PRD1, a plasmid-dependent broad host range DNA bacteriophage. J Virol. 1974 Sep;14(3):689–699. doi: 10.1128/jvi.14.3.689-699.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakula T. M., Savilahti H., Bamford D. H. Comparison of the amino acid sequence of the lytic enzyme from broad-host-range bacteriophage PRD1 with sequences of other cell-wall-peptidoglycan lytic enzymes. Eur J Biochem. 1989 Mar 1;180(1):149–152. doi: 10.1111/j.1432-1033.1989.tb14625.x. [DOI] [PubMed] [Google Scholar]
- Platt T. Transcription termination and the regulation of gene expression. Annu Rev Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- Reilly B. E., Nelson R. A., Anderson D. L. Morphogenesis of bacteriophage phi 29 of Bacillus subtilis: mapping and functional analysis of the head fiber gene. J Virol. 1977 Oct;24(1):363–377. doi: 10.1128/jvi.24.1.363-377.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savilahti H., Bamford D. H. Linear DNA replication: inverted terminal repeats of five closely related Escherichia coli bacteriophages. Gene. 1986;49(2):199–205. doi: 10.1016/0378-1119(86)90280-5. [DOI] [PubMed] [Google Scholar]
- Savilahti H., Bamford D. H. The complete nucleotide sequence of the left very early region of Escherichia coli bacteriophage PRD1 coding for the terminal protein and the DNA polymerase. Gene. 1987;57(1):121–130. doi: 10.1016/0378-1119(87)90183-1. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S. K., Ito J. Protein-primed replication of bacteriophage PRD1 genome in vitro. Virology. 1989 Jun;170(2):442–449. doi: 10.1016/0042-6822(89)90435-2. [DOI] [PubMed] [Google Scholar]