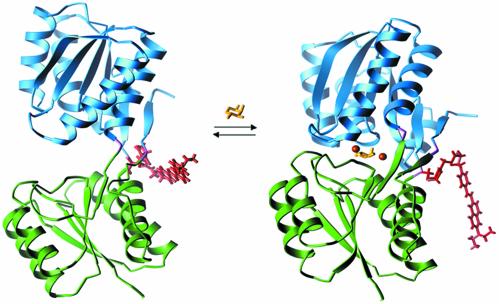
Fig. 1.
Ligand-mediated conformational changes in RBP. On binding of ribose the protein switches from an open conformation (Left; Protein Data Bank ID code 1URP; ref. 36) to a closed conformation (Right; Protein Data Bank ID code 2DRI; ref. 37). The conformational interchange can be monitored by covalent coupling of a thiol-reactive fluorophore to Cys-236 (31). A model of the conjugated styryl dye JPW4042 in each conformation was generated by identification of the minimum energy dye conformation (based on an in vacuo calculation using a semiempirical force field). The differences in the two conformations of the dye are primarily due to changes in the interactions between the dye and protein near the point of attachment. Also indicated are the positions of the Zn2+ ions in the three designed sites relative to the ribose in the closed conformation. Colors indicate the domain structure, fluorescent conjugate, and ligands of the receptor: green, N-terminal domain; blue, C-terminal domain; purple, hinge; yellow, ribose; red, fluorophore JPW4042; orange, zinc sites A (Right) and B (designs B1 and B2; Left).