Abstract
We estimated the amount of hepatocyte turnover in the livers of three woodchucks undergoing clearance of a transient woodchuck hepatitis infection by determining the fate of integrated viral DNA as a genetic marker of the infected cell population. Integrated viral DNA was found to persist in liver tissue from recovered animals at essentially undiminished levels of 1 viral genome per 1,000–3,000 liver cells, suggesting that the hepatocytes in the recovered liver were derived primarily from the infected cell population. We determined the single and multicopy distribution of distinct viral cell junctions isolated from small pieces of liver after clearance of the infection to determine the cumulative amount of hepatocyte proliferation that had occurred during recovery. We estimated that proliferation was equivalent to a minimum of 0.7–1 complete random turnovers of the hepatocyte population of the liver. Our results indicated that during resolution of the transient infections a large fraction of the infected hepatocyte population was killed and replaced by hepatocyte cell division.
Hepadnaviruses are small, enveloped DNA viruses that replicate by transcription of RNA pregenomes and reverse transcription to produce the DNA genomes that are found in the virus particles secreted from infected hepatocytes (1). Infected hepatocytes contain a large number of nascent viral DNA intermediates in the cytoplasm, in the form of nucleocapsids, and a small number of complete DNA genomes in the nucleus, in the form of double-stranded covalently closed circular DNA (cccDNA) (2, 3). The major types of hepadnaviruses that are studied include the human hepatitis B virus (HBV) and two animal virus models, the woodchuck hepatitis virus (WHV) (4) and the duck hepatitis B virus (5). All of these viruses share many structural, genetic, and biological traits.
Infection with hepadnaviruses can result in a number of different outcomes ranging from a subclinical transient infection to chronic infection resulting in a high risk for liver disease, primarily cirrhosis and hepatocellular carcinoma (6). Virus infection of hepatocytes is not normally cytopathic and therefore, in chronic infections, particularly those not associated with disease, it is common for all hepatocytes to be persistently and productively infected. Under these circumstances, high levels of infectious virus particles and viral antigens accumulate in the blood in excess of antibodies produced against the secreted proteins, and cellular immune responses are weak and unable to clear the infection from the liver (7). High levels of infection in the liver do not automatically result in chronic infections; however, many documented examples of transient infections also involve the total hepatocyte population and the accumulation of equivalent high levels of virus and antigens in the blood before the infection is resolved (8–13). It is striking that such widespread infection may be cleared from the liver without the occurrence of a life-threatening crisis of liver destruction, and this observation led to the early speculation that suppression of virus replication in infected hepatocytes, rather than hepatocyte death, is responsible for viral clearance (14).
Support for this notion comes from experiments with HBV transgenic mice, which replicate HBV in hepatocytes from a transgene that encodes the HBV pregenome (15). In these mice, HBV replication is exquisitely sensitive to the inflammatory cytokines IFN-γ and tumor necrosis factor α, responding with a strong reduction of viral replication in all hepatocytes within 24–48 h of inoculation of virus-specific cytotoxic T lymphocytes (16). Inflammation induced by a variety of agents, including adenovirus or leukochoriomeningitis virus infection, poly I:C, or IL-12 injection, produces a similar synchronous and rapid effect on all virus-producing hepatocytes (17–20), suggesting that the major effect of inflammation is a “bystander” effect of cytokines that eliminates HBV replication globally and noncytolytically in the liver surrounding the reactive cytotoxic T lymphocytes and recruited inflammatory cells. In contrast, resolution of natural infections in transiently infected ducks, woodchucks, and chimpanzees has been shown to occur by progressive elimination of virus-infected hepatocytes over a period of weeks, with a corresponding accumulation of uninfected cells (8–12). This piecemeal elimination of infected cells and accumulation of uninfected cells suggest that clearance occurs by killing of individual infected cells by virus-specific cytotoxic T lymphocytes, followed by the outgrowth of uninfected cells.
In recent years it has been vigorously debated how much hepatocyte destruction actually occurs during the resolution of a transient infection (10, 12, 13, 21). This question is unresolved for any individual case because methods for observing cell death in the liver are currently limited to histological examination of liver biopsies, from which only estimates of relative, but not absolute, rates of hepatocyte destruction at the point of tissue sampling can be made. In this study, we have obtained data on the cumulative amount of cell destruction during the resolution of hepadnavirus infection of woodchucks. We have used the presence of viral DNA sequences uniquely integrated into the DNA of a small fraction of infected hepatocytes as genetic markers of the population of infected cells to follow the fate of the infected cells during viral clearance. We found that after recovery from the infection there was no discernible reduction in the number of copies of integrated viral DNA in the liver, indicating that the uninfected cells of the recovered liver were derived primarily from infected cells. Examination of the copy number distribution of individual virus cell DNA junctions indicated that a significant degree of hepatocyte proliferation had occurred between the peak of infection and recovery, amounting to a minimum of 0.7–1 complete turnovers of the liver.
Materials and Methods
Transient Infection of Woodchucks with WHV. Adult woodchucks (Marmota monax) were housed in the Laboratory Animal Facility of the Fox Chase Cancer Center. All experiments carried out with these woodchucks were reviewed and approved by the center's Institutional Animal Care and Use Committee. At 7–8 months of age, three woodchucks were subjected to liver wedge biopsy as described (9, 22), and, 4 weeks later, they were inoculated i.v. with 2 ml of WHV-positive serum from a chronically infected woodchuck (WHV titer, ≈109 per ml). A control woodchuck was inoculated with 2 ml of PBS. Serum was collected weekly, and liver biopsy samples were collected at 4, 8, 12, and 16 weeks postinoculation (p.i.). Aliquots were stored at –80°C until use. Portions of each liver biopsy were also fixed with formalin, for histological analysis after staining with hematoxylin and eosin or ethanol/acetic acid (3:1) (9) for immunohistochemistry and in situ hybridization.
Nucleic Acid Analyses of Woodchuck Serum. Viral titers were determined by Southern blot hybridization assays (23, 24). Fifty microliters of woodchuck serum was layered on a 10–20% sucrose step-gradient containing 0.15 M NaCl and 20 mM Tris·HCl (pH 7.5). Virus was collected by centrifugation for 3 h at 50,000 rpm and 4°C in a Beckman SW 60 rotor, and the pellet was digested with 30 μl of 0.01 M Tris·HCl (pH 7.4), 0.01 M EDTA, 0.2% (wt/vol) SDS, and 1 mg/ml Pronase for 1 h at 37°C. The mixture was then subjected to electrophoresis into a 1.5% agarose gel and subsequent Southern blotting to nitrocellulose sheets. A 32P-labeled DNA probe representing the complete WHV genome was used for hybridization. Signals were quantified by using a Fuji phosphorimager, and virus titers were estimated by reference to a WHV DNA standard.
Sorbital Dehydrogenase (SDH) Assay. SDH levels in woodchuck serum, an indicator of liver injury (25), were determined by Ani-lytics (Gaithersburg, MD). Concentrations are expressed in international units per liter.
Immunohistochemistry and in Situ Hybridization. Immunoperoxidase assays for detection of WHV core antigen, proliferating cell nuclear antigen (PCNA), and CD3 were carried out on acetic acid/ethanol-fixed and paraffin-embedded tissues as described (10). Woodchuck CD3 was detected by using a rabbit polyclonal anti-human CD3 epsilon chain antiserum (DAKO), and PCNA was detected with mouse monoclonal anti-PCNA antibodies (DAKO). WHV core antigen was detected by using an antiserum raised in rabbits to purified recombinant WHV core antigen (10). In situ hybridization for WHV nucleic acids was performed on ethanol/acetic acid-fixed tissues by use of a nonstrand-specific digoxygenin-dUTP-labeled DNA probe representing the complete WHV genome (26, 27).
Extraction of Viral and Cellular DNA from Liver. For analysis of replicating and cccDNAs, liver tissue (≈20–50 mg) was homogenized in 1.5 ml of TE (10 mM Tris·HCl, pH 7.5/1 mM NaEDTA). For extraction of replicative intermediates, half the homogenate was adjusted to a total volume of 6 ml with 0.025 M Tris·HCl (pH 7.4), 0.01 M EDTA, 0.25% (wt/vol) SDS, 0.05 M NaCl, and 2 mg/ml Pronase and incubated for 1 h at 37°C. Nucleic acids were then extracted with a 1:1 mixture of phenol/chloroform and collected by ethanol precipitation. cccDNA was extracted as described (28). Briefly, the other half of the homogenate was adjusted to a volume of 3 ml with 0.01 M Tris·HCl (pH 7.5) and 0.01 M EDTA. Two hundred microliters of 10% (wt/vol) SDS was added, the mixture was briefly vortex mixed, and 1 ml of 2.5 M KCl was added. After incubation at room temperature for 20 min, potassium dodecyl sulfate protein complexes were collected by centrifugation at 10,000 rpm for 20 min in a Sorvall SS34 rotor at 4°C. The supernatant was subjected to phenol/chloroform extraction. Nucleic acids were precipitated with ethanol overnight at room temperature. Southern blot hybridization analysis was then carried out as described (9). To quantify relative amounts of replicative intermediates, the amount of cell DNA was determined by fluorimetry (9). To quantify cccDNA, DNA extracted from a predetermined number of cells was assayed, as assessed by counting in a hemocytometer of fluorescent nuclei in the initial cell homogenates after staining with ethidium bromide.
For quantification of integrated viral DNA, the potassium dodecyl sulfate pellet was suspended in 0.4 ml of TE (10 mM Tris·HCl, pH 7.5/1 mM NaEDTA) and dissolved by incubation at 55°C with frequent vortex mixing. The solution was adjusted to 50 mM with NaCl and extracted two times with an equal volume of phenol and precipitated with ethanol. DNA concentrations were determined by fluorimetry with SYBR Green dye.
For the isolation of cellular DNA from small liver fragments, frozen liver (≈1 mg) was homogenized in 0.5 ml of TE at 0°C, and 0.01 ml was removed for counting the total number of nuclei. The remaining sample was adjusted to 0.2 M NaCl by the addition of 0.4 ml of 0.4 M NaCl and incubation at 0°C for 15 min. Nuclei were pelleted by microfuge centrifugation, and the supernatant fluid was discarded. The pellet was resuspended in TE, adjusted to 0.2% Triton X-100, 0.2% SDS, and 500 μg/ml proteinase K. The sample was incubated for 2 h at 55°C with frequent vortex mixing and then overnight at 37°C, adjusted to 50 mM NaCl, extracted twice with phenol/chloroform (1:1 vol/vol), ethanol-precipitated, and dissolved in TE.
PCR Primers and Linkers. A nonphosphorylated linker was designed to ligate unidirectionally to blunt ends of cellular DNA generated by PvuII so that the recognition site would be regenerated. The strand intended to ligate to the 5′ phosphate of the PvuII site, LNK-D, consisted of the sequence 5′-TGCCCTCGTCCCTAATGCAG, and this oligonucleotide was also used as the linker-specific primer in amplification of viral cell junctions. The complementary strand of the linker was 5′-CTGCATTAGGGA. The resulting partial duplex of the double-stranded linker contained one blunt end with a PvuII half site (.CAG-3′) capable of ligating to the 5′-P-CTG. of the PvuII blunt ends of cellular DNA. At the other end of the linker, the 3′ OH was recessed and unable to ligate to a PvuII blunt end.
The viral-specific primer, WHV2250–2224, was used with the linker-specific primer, LNK-D, to amplify viral cell junctions. This oligonucleotide primed DNA synthesis in the minus direction on the WHV genome, toward the left end limits of viral cell junctions, into cellular DNA toward the ligated linker. The S1 primer set consisted of primers WHV2185–2167 and WHV2195–2216, and the nested S2 primer set consisted of primers WHV2165–2145 and WHV2217–2245. All viral-specific primers are designated by WHV followed by the inclusive nucleotides (5′-3′ direction) numbered according to the sequence of Galibert et al. (29).
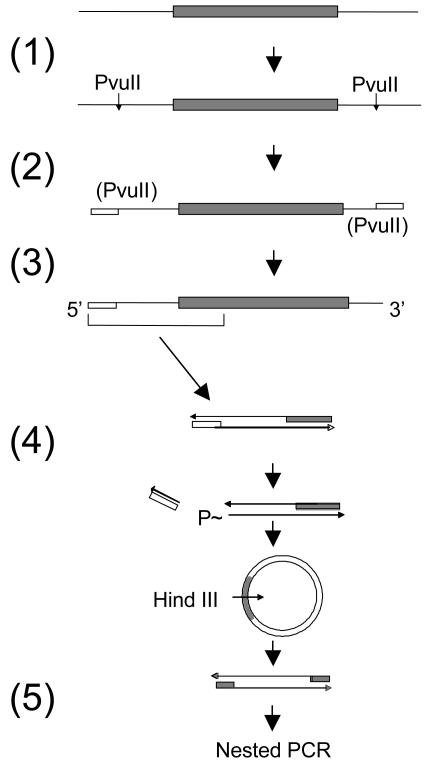
Assays for Integrated DNA. The standard quantitative assay for integrated DNA measured the detection of viral cell junctions from a known number of cells, calculated from the amount of cellular DNA used in the assay. Specifically, we detected the left end junctions, which are those in which cellular sequences were joined to the left end of a viral DNA sequence oriented left to right by increasing nucleotide number. Left end junctions are clustered downstream of nucleotide position 1935 (see Supporting Text, which is published as supporting information on the PNAS web site, www.pnas.org). Viral cell junctions were detected by a procedure that involved sequential application of ligation-mediated PCR and inverse nested PCR. The steps are outlined in Fig. 1.
Fig. 1.
Amplification of viral cell junctions. Viral DNA (shaded box) is schematically shown integrated into cell DNA (solid line). Steps are indicated as follows and are described in detail in Materials and Methods: 1, digestion of cellular PvuII sites; 2, ligation of ends to LNK-D (open box) and termination of 3′ ends (data not shown); 3, denaturation and endpoint dilution amplification of one strand with viral and linker-specific primers; 4, removal of LNK-D sequences with PvuII, circularization, and digestion with HindIII; and 5, nested PCR with viral-specific primers.
PvuII digestion. Cellular DNA (≈2 μg) was digested with 2 units of PvuII, and the DNA was recovered with a QIAquick Spin Column (Qiagen, Valencia, CA) and eluted in 50 μl of elution buffer (10 mM Tris·HCl, pH 8.5).
Linker ligation. The DNA was adjusted to 50 mM Tris·HCl (pH 7.5), 10 mM MgCl2, 1 mM DTT, and 0.01% Nonidet P-40 (ligase buffer). The unphosphorylated oligonucleotide linker, LNK-D, (100 pmol) was added along with 100 units of T4 DNA ligase and the linker (+) strand ligated to the 5′ phosphorylated ends of the PvuII sites by incubation at 25°C for 2 h. Termination of all 3′ ends was performed after the addition of a mixture of dideoxynucleoside triphosphates at 50 μM each and 5 units of TaqDNA polymerase and incubation at 72°C for 1 h (data not shown). This step was carried out to reduce the amplification of cell DNA containing a LNK-D sequence at both ends. The DNA was recovered, freed of excess linker and dideoxynucleoside triphosphates by using a QIAquick Spin Column, and eluted in 50 μl of elution buffer. Linker-ligated DNA was diluted and subjected to PCR.
First PCR. DNA was denatured at 95°C for 2 min and added to a PCR mixture containing 25 mM MgCl2, 100 nM LNK-D primer, 200 nM primer 1 (2250–2217), 25 units/ml AmpliTaq Gold DNA polymerase, and AmpliTaq Gold buffer II. Aliquots (10 μl) were distributed into the wells of a PCR microplate (Fisher) and subjected to 10 min at 95°C, 45 cycles of 20 sec at 95°C, 20 sec at 58°C, and 3 min at 72°C, followed by 10 min at 72°C. Templates amplified by this regimen consisted of viral cell junctions and a residual background of PvuII fragments of cellular DNA with LNK-D primer binding sites at both ends. Individual viral cell junctions were amplified from this complex mixture of products in each well by inverse nested PCR.
Circularization. Approximately 0.4 μl from each well was transferred to a second PCR microplate with the aid of a 96-pin replicator (Nalge Nunc International). The wells of the second microplate each contained 5 μl of ligase buffer containing 2,000 units/ml T4 DNA ligase, 20 units/ml PvuII, and 10 units/ml exonuclease I. The plate was incubated for 30 min at 37°C, then for 2 h at 25°C. In this step, viral cell junctions were circularized by the following reactions: (i) removal of the LNK-D sequences by cleavage of the reconstituted PvuII site, producing a 5′ phosphate, and (ii) removal of the protruding 3′ nucleotide from the other end of the amplified viral cell junction that results from nontemplated addition by the polymerase, ligation of the resulting 3′ OH of the blunt end to the 5′ phosphate to produce a circular double strand in which the ligated strand is covalently closed.
Nested PCR. The viral cell junctions were then linearized at the HindIII site in the viral sequences and amplified specifically by nested primers specific for the viral sequences. Approximately 0.4 ml from each well of the ligation plate was transferred to a third PCR microplate whose wells each contained 10 μl of the PCR mix consisting of Herculase Hotstart DNA Polymerase (25 units/ml, from Stratagene), Herculase buffer supplemented to contain 2.5 mM MgCl2, 200 μM each dNTP, 100 nM each S1 primer set, and 100 units/ml HindIII. The plate was incubated at 37°C and then subjected to 3 min at 95°C, 30 cycles of 20 sec at 95°C, 20 sec at 54°C, and 3 min at 72°C, followed by 3 min at 72°C.
Finally, ≈0.1 μl from each well was transferred to a fourth PCR microplate whose wells each contained 10 μl of the Herculase PCR mix above, but with 100 nM each of the nested S2 primer set, and omitting the HindIII. This plate was subjected to the same cycling conditions as above but without the 37°C prior incubation.
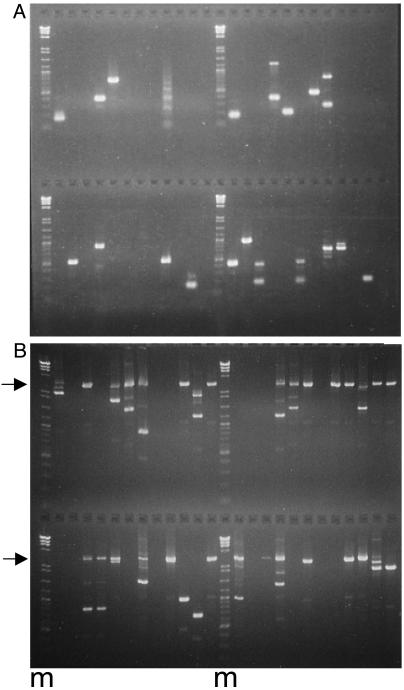
The entirety of each well was mixed with gel loading buffer and analyzed by agarose gel electrophoresis (1.3% LE agarose, FMC Bioproducts) in a Tris-acetate electrode buffer to visualize the DNA products from single template molecules. The starting amount of LNK-D ligated DNA was adjusted by pilot experiments so that half or fewer of the wells of the PCR plate contained products, thereby favoring unique products in each positive well (see example in Fig. 3A). The DNA bands were excised from the gel, and the DNA was isolated with a QIAEX II reagent kit procedure (Qiagen) modified for a 200-μl PCR microplate. DNA products were subjected to automated sequencing and analyzed individually for viral sequences by using the blast2 sequences utility of the National Center for Biotechnology Information web site. Sequencing was necessary to distinguish viral cell junctions from pieces of viral cccDNA that were amplified because of cleavage and linker ligation at a PvuII star site and to map the recombination site. In liver samples from woodchucks collected during the replication phase, the viral cell junctions represented 20–50% of the PCR products, whereas, after recovery, >95% of the recovered products were viral cell junctions.
Fig. 3.
Endpoint dilution assays for viral cell junctions and cccDNA. Agarose gel (1.3%) electrophoresis of PCR assays with endpoint dilutions of templates. Each reaction was loaded in a single lane. (A) Single viral cell junctions were amplified in 48 separate reactions as depicted in Fig. 1. Individual bands were identified after extraction from the gel and sequencing. (B) cccDNA from a 16-week biopsy of WC380, digested with HindIII, adjusted to limiting dilution, and amplified by nested PCR in 48 separate reactions. The arrows mark the migration position of WT (3 kb) DNA, whereas defective molecules migrated more rapidly. The number of defective molecules was taken to be equal to the number of products <3 kb, whereas the number of WT molecules was estimated from the Poisson distribution by the number of lanes (reactions) that did not contain a 3-kbp PCR product. Markers (m) were a mixture of a HindIII digest of bacteriophage lambda DNA and a 100-bp ladder (New England Biolabs).
Assay for Copy Number Distribution of Viral Cell Junctions. A simplified procedure to amplify all of the viral cell junctions from small pieces of liver for the estimate of proliferation was used, because these samples collected at 16 weeks postinfection contained much less interfering cccDNA. This procedure used inverse nested PCR. Cellular DNA was digested with SacI, which cuts at a single site in the viral sequence. The SacI was inactivated by heating at 80°C for 15 min, and the DNA was diluted to a concentration to favor intramolecular ligation of ≈4 μg/ml in ligase buffer containing 400 units/ml T4 DNA ligase and incubated at 25°C for 2 h. The DNA was recovered from the ligation with a QIAquick Spin Column and eluted with 100 μl of elution buffer. The DNA solution was adjusted with restriction enzyme buffer II (New England Biolabs), circularized molecules were linearized with HindIII (100 units/ml), and molecules derived from cccDNA were removed as possible templates by digestion with AflII. The remaining viral cell junctions were diluted and amplified in PCR microplates by using the S1 and nested S2 set of primers, and individual products were isolated and sequenced as described above. About 50% of the products were derived from viral cell junctions, and the rest were from small, defective cccDNA molecules in which the AflII site had been deleted.
Assay for Defective cccDNA. Extracted cccDNA was linearized and diluted so that ≈50 molecules would be distributed in 96 10-μl reactions in a PCR microplate. Individual cccDNA molecules were amplified for 30 cycles by using the Herculase Hotstart conditions as described above by primer set S1 but with a 5-min elongation step, followed by transfer to a second PCR plate containing reaction mixes with nested primer set S2 for an additional 30 cycles of amplification. The products were analyzed by agarose gel electrophoresis to detect products derived from cccDNAs with discernible deletions. An example of such an assay is shown in Fig. 3B.
Results
Course of the Infection. Table 1 compares the virus levels observed in the blood of the three woodchucks over the study period. The viremia in each animal appeared after inoculation in a transient manner, accompanied by a transient but delayed increase in serum SDH, an indicator of liver cell lysis. Increases in SDH levels distinctly followed the reduction in viremia (Table 1). The course of the infection was unremarkable compared with previous woodchuck inoculation experiments (9–11) and resulted in a total infection of the liver followed by a nearly complete clearance of markers of virus replication.
Table 1. Serum assays for WHV DNA and SDH.
| Weeks p.i.
|
WHV DNA viremia, 10-7 per ml*
|
SDH, units†
|
||||
|---|---|---|---|---|---|---|
| WC377 | WC378 | WC380 | WC377 | WC378 | WC380 | |
| 0 | <1 | <1 | <1 | 7 | 6 | 5 |
| 1 | <1 | |||||
| 2 | 1.5 | <1 | 7 | 15 | ||
| 3 | 15 | 200 | 20 | |||
| 5 | 20 | 500 | 26 | 64 | ||
| 6 | 35 | 90 | 20 | 28 | 127 | 40 |
| 7 | 160 | 50 | 34 | 75 | 31 | |
| 8 | 110 | 20 | 40 | 21 | 72 | |
| 9 | 50 | 1 | 70 | 42 | 189 | |
| 11 | <1 | <1 | 10 | 146 | 191 | 162 |
| 12 | <1 | 183 | 53 | 125 | ||
| 14 | 32 | 37 | 31 | |||
| 15 | 31 | 40 | 35 | |||
| 16 | 31 | 31 | 32 | |||
WHV DNA levels were assayed by gel electrophoresis and Southern blot hybridization assay of virus particles pelleted from 50 μl of serum.
SDH levels in the uninfected control during the same period ranged between 4 and 38 units.
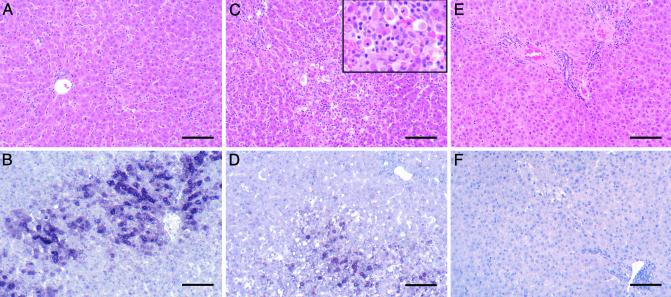
Histological Measurements. Fig. 2 shows the histological appearance and in situ hybridization of the livers of woodchucks WC377, WC378, and WC380 during the phase in which viral infection in the liver was being cleared. Table 2 shows the measurement of several histological and virological markers in the livers over the course of the experiment obtained by analysis of all of the biopsies. The time course of WHV infection and accumulation of infected cells and viral DNA (both cccDNA and replicative intermediates) in the livers occurred with similar kinetics in each animal, with peak levels of infection being found in the 8-week biopsies, and almost all evidence of virus replication disappearing by either 12 or 16 weeks p.i. Both in situ hybridization to detect WHV nucleic acids (Table 2 and Fig. 2) and immunohistochemical staining for viral core antigens (data not shown) indicated that >95% of all hepatocytes were infected in all animals at 4 and 8 weeks p.i. Peak infiltration of the livers with CD3+ mononuclear cells (lymphocytes) followed closely behind peak viral DNA levels occurring at either 8 or 12 weeks p.i. For two woodchucks (WC377 and WC380), the greatest elevation in CD3+ cells was detected in biopsies collected when the number of WHV-positive hepatocytes had decreased and when infection was already partially cleared from the liver (12 weeks). CD3+ cells were prominent in the periportal areas and throughout the lobules where the number of CD3+ cells rose to 24–40% of the total number of hepatocytes. Maximum apoptotic indices varied more among the animals than other histological markers, reaching values of up to 10% of the hepatocytes in WC377 at 12 weeks, but peak values were as low as 1.4% and 1.6% in the other two infected animals. Apoptotic bodies were often positive for in situ hybridization (Fig. 2D) and core antigen staining (data not shown). PCNA staining of cell nuclei showed evidence of regenerative hepatocyte proliferation, with peak levels between 0.8% and 1.6% of hepatocytes. The timing of changes in each of these markers, i.e., a decrease in the percentage of infected cells and viremia and increases in CD3+ cells, apoptotic counts, and PCNA staining, all coincided with increases in levels of SDH in the serum (Table 1), indicating hepatocyte death during clearance of infection. Thus, the overall picture of the resolution phase of infection was that of acute inflammation and hepatocyte injury that was most active at the end of the period of peak liver infection and viremia at 8 or 12 weeks p.i. Because the intensity of these histological changes is related to the rate of cell damage, it is expected that such changes would be most severe during periods in which a large number of infected hepatocytes were present and being acted on as targets of immune effectors. However, although it was clear that hepatocyte death was occurring, we found no basis in any of these measurements for estimating the cumulative amount of hepatocyte turnover associated with viral clearance.
Fig. 2.
Histological appearance and in situ hybridization of the liver at 12 weeks p.i. Liver biopsy samples collected from woodchucks WC380 (A and B), WC377 (C and D), and WC378 (E and F) at 12 weeks p.i. were stained with hematoxylin and eosin (A, C, and E) to detect histological changes or subjected to in situ hybridization and counterstained with hematoxylin (B, D, and F) to detect WHV nucleic acids. Liver of WC380 showing infiltration of mononuclear cells, apoptotic hepatocytes (A), and WHV nucleic acids in 49% hepatocytes (B). Liver of WC377 showing marked inflammation, apoptotic hepatocytes (Inset) that contributed up to 10% of the total hepatocyte population (C), and WHV nucleic acids in 24% of hepatocytes, some with features of apoptosis (D). Liver of woodchuck WC378 at 12 weeks p.i. showing essentially normal liver histology (E) and no detectable WHV nucleic acids (F). (Final magnification, ×84; Inset, ×210; bars = 100 μM.)
Table 2. Virological and histological markers in liver biopsies.
| Virological markers
|
Histological markers
|
||||||
|---|---|---|---|---|---|---|---|
| Animal | Weeks p.i. | ISH* | cccDNA† | RI† | CD3‡ | Apoptosis§ | PCNA¶ |
| WC377 | -4 | 0 | 0 | 0 | 3 | 6 | 40 |
| 4 | >95 | 11 | 280 | 3 | 8 | 20 | |
| 8 | >95 | 33 | 1,700 | 16 | 59 | 70 | |
| 12 | 24 | 1 | 22 | 40 | 1,000 | 160 | |
| 16 | <0.01 | <1 | <10 | 5 | 2 | 7 | |
| WC378 | -4 | 0 | 0 | 0 | 2 | 3 | 2 |
| 4 | >95 | 12 | 600 | 3 | 22 | 1 | |
| 8 | >95 | 28 | 1,600 | 36 | 158 | 120 | |
| 12 | <0.01 | <1 | <10 | 13 | 50 | 10 | |
| 16 | <0.01 | <1 | <10 | 11 | 4 | 160 | |
| WC380 | -4 | 0 | 0 | 0 | 1 | 18 | 7 |
| 4 | >95 | 6 | 120 | 3 | 5 | 1 | |
| 8 | >95 | 61 | 720 | 17 | 34 | 1 | |
| 12 | 49 | 13 | 400 | 24 | 140 | 80 | |
| 16 | <0.01 | <1 | <10 | 5 | 12 | 7 | |
In situ hybridization for viral nucleic acids, percentage of hepatocytes positive.
Molecules per total liver cells by Southern blot assay; RI, replicative intermediates.
Positive cells per 100 hepatocytes (fields containing 3,500-7,700 hepatocytes scored).
Apoptotic hepatocytes per 10,000 hepatocytes (1,080-5,000 hepatocytes scored).
Positive hepatocytes per 10,000 hepatocytes (5,440-20,520 hepatocytes scored).
Integrated DNA Levels During the Course of the Infection. To obtain information on the cumulative turnover of the population of infected hepatocytes, we studied a property expected to act as a unique stable genetic marker of representative infected cell lineages, i.e., the integrated viral DNA molecules that are found in a small fraction of infected cells (30). Individual left end viral cell junctions of integrated DNAs were amplified from all biopsy specimens (e.g., Fig. 3A) and identified by sequencing. The frequency of occurrence of integrated DNA was estimated from the number of viral cell junctions that were identified and the amount of cellular DNA analyzed, assuming that each diploid cell contained a nominal 10 pg of DNA. The results of these analyses are shown in Table 3. It is evident that although cccDNA levels decreased by several orders of magnitude during the resolution of infection (Tables 2 and 4), there was no discernible decrease in viral cell junctions. This result suggests that the recovered liver consisted primarily of cells derived from the infected cell population.
Table 3. Integrated WHV DNA in infected liver.
| Animal | Weeks p.i. | DNA, ng* | V C junctions† | Frequency‡ |
|---|---|---|---|---|
| WC377 | -4 | 188 | 0 | <1 |
| 4 | 140 | 4 | 3 | |
| 8 | 118 | 15 | 13 | |
| 12 | 116 | 10 | 9 | |
| 16 | 78 | 8 | 10 | |
| WC378 | -4 | 182 | 0 | <1 |
| 4 | 134 | 15 | 11 | |
| 8 | 116 | 8 | 7 | |
| 12 | 98 | 4 | 4 | |
| 16 | 232 | 9 | 4 | |
| WC380 | -4 | 92 | 0 | <1 |
| 8 | 157 | 13 | 8 | |
| 12 | 165 | 21 | 13 | |
| 16 | 126 | 8 | 6 |
Total cellular DNA analyzed.
No. of viral cell junctions identified.
No. of viral cell junctions per 10,000 cells, assuming each cell contained 10 pg of total DNA.
Table 4. Enrichment of defective cccDNA after clearance.
| Assayed*
|
cccDNA per cell†
|
||||||
|---|---|---|---|---|---|---|---|
| Animal | Weeks p.i. | Cells | WT | Defective | WT | Defective | Fraction Defective |
| WC377 | 4 | 20 | 94 | 2 | 4.7 | 0.1 | 0.02 |
| 16 | 6,300 | 17 | 9 | 0.0027 | 0.0014 | 0.34 | |
| WC378 | 4 | 9 | 61 | 4 | 6.6 | 0.4 | 0.04 |
| 16 | 10,800 | 31 | 27 | 0.0029 | 0.0025 | 0.46 | |
| WC380 | 4 | 44 | 115 | 3 | 2.6 | 0.07 | 0.03 |
| 16 | 36,400 | 90 | 52 | 0.0025 | 0.0014 | 0.36 | |
cccDNA from a known number of cells was analyzed. The number of PCR products migrating at 3 kbp (WT) and the number migrating at <3 kbp (defective) are shown.
Calculated.
Proliferation of Infected Cells Containing Integrated WHV DNA During Resolution of the Infection. As previously reported for duck hepatitis B virus (31), the viral sites of joining of viral DNA to nonviral (cell) DNA were clustered in a region corresponding to the left ends of linear DNA forms (see Fig. 4, which is published as supporting information on the PNAS web site). This distribution of recombination sites is consistent with a nonhomologous end joining reaction between the left end of linear DNA and a cellular DNA end. To determine whether viral cell junctions in infected hepatocytes could serve as unique markers of hepatocyte lineage, we examined the sequences of 127 viral cell junctions identified in 4- and 8-week biopsy samples of infected liver (Table 3 and data not shown) for the appearance of repeated copies of the same sequence. Of 127 viral cell junctions identified, all but 2 were unique (the repeated junctions were found in the same biopsy), indicating that integration of viral DNA did not occur at highly preferred sites. For this reason we assumed that the occurrence of multiple copies of the same virus cell junction would have been derived from the division of a cell that contained that particular junction. To estimate the amount of cell division that occurred in the population of infected cells during the resolution of infection we extracted DNA from small pieces of the liver containing 105 to 106 cells and amplified and sequenced all of the viral cell junctions contained therein. The population of sequences was then examined for the occurrence of unique vs. repeated copies. A summary of these data are shown in Table 5.
Table 5. Complexity of viral cell junctions.
| Copy no.†
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Animal | Sample, weeks | Total V C joints* | 1 | 2 | 3 | 4 | >4 | Total clones‡ | Complexity |
| Mixed pool§ | 4,8 | 127 | 125 | 1 | 0 | 0 | 0 | 126 | 0.99 |
| WC377 | 16 | 51 | 18 | 3 | 2 | 1 | 2 | 26 | 0.5 |
| WC378 | 16 | 26 | 7 | 4 | 0 | 1 | 1 | 13 | 0.5 |
| WC380 | 16 | 52 | 19 | 5 | 3 | 2 | 1 | 30 | 0.6 |
No. of viral cell junctions recovered and sequenced.
No. of clones containing the indicated copy number.
Sum of previous five columns.
Viral cell junctions from 4- and 8-week biopsies of all animals.
Evidence for cell proliferation, i.e., multicopy junctions, can be seen in all three postrecovery liver samples. The data were used to calculate the “complexity” of the population of viral cell junctions, i.e., the number of different sequences detected in the sample divided by the total number of junctions detected. The complexity of the population of cells with integrated DNA was 0.5, 0.5, and 0.6 in the three woodchucks, where a complexity of 1.0 would correspond to a sample with no multicopy junctions. A complexity of <1.0 is direct evidence of hepatocyte proliferation, induced presumably by an equivalent amount of cell death.
The presumed turnover giving rise to the observed complexity was related quantitatively to that expected from a process of random cell turnover. Using a model (see Supporting Text) that assumes a starting cell population size corresponding to the total number of viral cell junctions, we calculated the effect of sequential random cell killing and division cycles on the distribution of multicopy junctions and the complexity of the population of junctions after different amounts of turnover. From this model, we estimated that the reduced complexity of the postrecovery samples (0.5, 0.5, and 0.6) corresponded to 1, 1, and 0.7, respectively, complete turnovers of the population of infected cells. Incorporating into this model the effect of the efficiency of detection of viral cell junctions (Table 6, which is published as supporting information on the PNAS web site), we estimated the turnover expected to give rise to the reduced complexity, assuming an efficiency of detection of 50%, as 2, 2, and 1.4 complete turnovers of the population of infected cells.
Selection of Defective Viral Genomes During cccDNA Clearance. Viral cccDNA in the nucleus of infected cells consists of a mixture of two types of molecule. The predominant type is derived from a precursor circular double-stranded DNA molecule that is generally a complete copy of the genome and active for expression of all viral proteins. A minor type of cccDNA is derived by nonhomologous end joining reactions between the ends of linear double-stranded DNA, resulting in gross deletions of parts of the viral genome extending in both directions from near the start site of pregenome transcription (32–34). Most such genomes are predicted to be defective in replication and protein expression and may be recognized by their reduced size. We observed that, after resolution of the infection in the livers of transiently infected woodchucks, the residual amount of cccDNA was greatly enriched in defective molecules.
Single copies of cccDNA molecules were amplified by nested PCR of samples diluted so that individual reactions contained an average of less than one molecule, and the PCR products were analyzed by agarose gel electrophoresis to identify products derived from grossly deleted cccDNA templates. An example of a collection of such products amplified from a 16-week sample is shown in Fig. 3B. Grossly deleted molecules whose mobility was shifted constituted up to one-half of all molecules in the 16-week sample but were infrequent in the 4-week biopsies. A summary of the data for the three woodchucks is seen in Table 4. It is clear that the reduction of full-length cccDNA molecules was substantially greater than the reduction in defective molecules. One explanation for this selective destruction of full-length cccDNA would be that cells containing full-length molecules were removed by an antigen-dependent process, according to their ability to produce the full array of viral proteins. An alternative explanation would be that cccDNA molecules were destroyed by a process that depended on their physical target size.
Discussion
The transient WHV infection seen after inoculation of three adult woodchucks followed the typical course described by us and others (9–11). Infection occurring in >95% of hepatocytes was cleared over a period of 4–8 weeks without any apparent life-threatening crisis and with levels of inflammation, necrosis, and apoptosis in two animals that was similar to that previously reported, whereas in one animal (WC377) five times higher levels of apoptosis were observed. Histological changes in the liver (data not shown) ranged from relatively mild (WC378) to moderate or severe (WC380 and WC377, respectively). Evidence for hepatocyte destruction, seen in biopsies from the clearance phase, consisted of necrosis, apoptosis, and regeneration. Viral markers in the liver, i.e., replicative intermediates, cccDNA, and infected cells, decreased approximately in parallel, as would be expected if infected cells were being removed primarily by killing during the inflammatory process. However, it was not apparent from the histological observations how to estimate the total amount of killing that occurred during the resolution phase of the infection.
To address this question, a quantitative assay for integrated DNA was developed to follow the fate of the infected cell population, using integrated DNA as a genetic marker specific to that population of cells. Had the infected cells been killed and replaced by uninfected stem cells, the integrated DNA characteristic of the infected cell population would have disappeared along with the infected cells. Because we did not observe such a gross disappearance of integrated DNA, we concluded that a substantial fraction of the cells of the recovered liver were derived from the population of infected cells. This result allowed us to estimate the amount of proliferation that these cells in the recovered liver had undergone during the recovery phase. Our estimate was derived from measurements of the distribution of unique and multicopy viral cell junctions within small samples of liver. The estimate depends on the validity of two assumptions. The first is that all of the progeny of a single dividing hepatocyte were found in the same immediate vicinity, in a cluster, and therefore that entire clusters were recovered in the small piece of liver analyzed. BrdUrd labeling experiments in the three woodchucks revealed that 42–85% of cells labeled after a single injection could be found as pairs in thin sections after 4 weeks (9). This result implies that multiple cell divisions would result in clustering of progeny. If such clusters did exist their dimensions would be only a fraction of the total dimensions of the piece of liver analyzed. For example, even a large clonal cluster of 100 hepatocytes would make up only 1/1,000 of the volume of a sample containing 105 cells, making it improbable that dissection of the liver tissue would result in dissection of the cluster.
A second assumption of our estimate of proliferation is that infected cells with integrated DNA behaved in all ways similar to infected cells without integrated DNA. At least, in the initial stages of resolution, antigen-dependent cell killing would be directed primarily by presentation of antigens produced as a result of virus replication rather than from integrated DNA, a minor template in the infected cell that may or may not produce sufficient antigenic peptides to be presented by the cell in competition with endogenous peptides. In addition, the target sites for integration are numerous, and we expect that integration into only a small fraction of these sites would affect cell growth or viability. This assumption implies that the population of cells with integrated DNA is representative of the whole population of infected cells.
Proliferation was likely in response to hepatocyte death. During the course of the infection each of the woodchucks actually declined in body mass (data not shown), arguing against normal growth and in favor of hepatocyte turnover as a source of proliferation. The amount of turnover estimated by this method depends on the efficiency with which any particular viral cell junction was detected. The turnover values of 0.7 and 1 were calculated by assuming that every type of viral cell junction that was detected in the assay was detected with 100% efficiency. It would be difficult to argue that we detected each viral cell junction with 100% efficiency, however, because failure to prepare and amplify a viral cell junction could occur at four different steps: extraction of cell DNA, circularization, recovery of templates, and amplification (assuming that all three restriction enzyme digestions were 100% complete). For example, although each step may be as much as 85% efficient, the combined efficiency of detection would be 52%, and the efficiency may be different for each viral cell junction. With this in mind, the estimated turnover must be considered to be the minimum estimate, but the actual turnover was likely to be higher.
Past lack of evidence of a high degree of hepatocyte turnover during clearance of infection has been used to support a significant role for noncytolytic “purging” of virus infection, including cccDNA, from individual cells (13, 35). Elimination of cccDNA from infected cells is crucial to cell curing because it is cccDNA that is the source of sustained virus production. Our analysis showed that cumulative hepatocyte turnover was extensive and similar in all three animals, irrespective of the histological evaluations. Therefore, consideration of an alternative mechanism must be revived (21), namely that a large fraction of infected hepatocytes was killed, and uninfected cells arose from the proliferation of remaining infected hepatocytes. This scenario requires that de novo infection of uninfected hepatocytes be prevented, a role that might be ascribed to the antiviral effects of inflammatory cytokines.
Studies of antiviral therapy with inhibitors of viral DNA synthesis in natural infections of woodchucks and ducks (36–40) have shown that inhibition of viral DNA synthesis alone can result in decreases in the number of infected hepatocytes, in the apparent absence of a role for the immune system. These reductions are thought to occur by elimination of infected hepatocytes through cell turnover and compensatory cell proliferation, leading to reduction of cccDNA copy number and eventual segregation and accumulation of uninfected cells. Significantly, in one study in duck hepatitis B virus-infected ducks treated with an antiviral drug that caused liver cell toxicity (41), the loss of infected hepatocytes was markedly accelerated, suggesting that cell death and proliferation are key components in virus elimination. Although cytokine-induced purging of virus from infected cells would be a truly noncytolytic process, the segregation and accumulation of uninfected hepatocytes through repeated cell division could not be considered noncytolytic because it is cell death that drives the proliferation. Our data do not distinguish between these two alternatives because both scenarios predict that integrated DNA would survive clearance, and both scenarios provide for hepatocyte turnover.
Thus, although resolution of the WHV infections was accompanied by an amount of death and regeneration equivalent to the entire liver or more, whether this turnover was essential for resolution of the infection even in the presence of antiviral cytokines is unclear. Our data argue, however, that the amount of killing and regeneration involved must have played a significant role in the direct elimination of infected cells (21).
Supplementary Material
Acknowledgments
We thank Darren Miller for technical support and Robert Lanford for a critical reading of the manuscript. A.R.J. was supported by a project grant from the National Health and Medical Research Council of Australia. J.S. and W.S.M. were supported by grants from the National Institutes of Health.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on May 1, 2001.
Abbreviations: cccDNA, covalently closed circular DNA; HBV, hepatitis B virus; WHV, woodchuck hepatitis virus; p.i., postinoculation; SDH, sorbital dehydrogenase; PCNA, proliferating cell nuclear antigen; TE, Tris/EDTA.
References
- 1.Summers, J. & Mason, W. S. (1982) Cell 29, 403–415. [DOI] [PubMed] [Google Scholar]
- 2.Tuttleman, J. S., Pugh, J. C. & Summers, J. W. (1986) J. Virol. 58, 17–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuttleman, J. S., Pourcel, C. & Summers, J. (1986) Cell 47, 451–460. [DOI] [PubMed] [Google Scholar]
- 4.Summers, J., Smolec, J. M. & Snyder, R. (1978) Proc. Natl. Acad. Sci. USA 75, 4533–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mason, W. S., Seal, G. & Summers, J. (1980) J. Virol. 36, 829–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ganem, D. & Schneider, R. J. (2001) in Fields Virology, eds. Fields, B. N., Howley, P. M., Griffin, D. E., Lamb, R. A., Martin, M. A., Roizman, B., Straus, S. E. & Knipe, D. M. (Lippincott Williams & Wilkins, Philadelphia), 4th Ed., Vol. 2, pp. 2923–2969. [Google Scholar]
- 7.Rehermann, B., Fowler, P., Sidney, J., Person, J., Redeker, A., Brown, M., Moss, B., Sette, A. & Chisari, F. V. (1995) J. Exp. Med. 181, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jilbert, A. R., Wu, T. T., England, J. M., Hall, P. M., Carp, N. Z., O'Connell, A. P. & Mason, W. S. (1992) J. Virol. 66, 1377–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kajino, K., Jilbert, A. R., Saputelli, J., Aldrich, C. E., Cullen, J. & Mason, W. S. (1994) J. Virol. 68, 5792–5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guo, J. T., Zhou, H., Liu, C., Aldrich, C., Saputelli, J., Whitaker, T., Barrasa, M. I., Mason, W. S. & Seeger, C. (2000) J. Virol. 74, 1495–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponzetto, A., Cote, P. J., Ford, E. C., Purcell, R. H. & Gerin, J. L. (1984) J. Virol. 52, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thimme, R., Wieland, S., Steiger, C., Ghrayeb, J., Reimann, K. A., Purcell, R. H. & Chisari, F. V. (2003) J. Virol. 77, 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guidotti, L. G., Rochford, R., Chung, J., Shapiro, M., Purcell, R. & Chisari, F. V. (1999) Science 284, 825–829. [DOI] [PubMed] [Google Scholar]
- 14.Edgington, T. S. & Chisari, F. V. (1975) Am. J. Med. Sci. 270, 212–227. [PubMed] [Google Scholar]
- 15.Guidotti, L. G., Matzke, B., Schaller, H. & Chisari, F. V. (1995) J. Virol. 69, 6158–6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guidotti, L. G., Ishikawa, T., Hobbs, M. V., Matzke, B., Schreiber, R. & Chisari, F. V. (1996) Immunity 4, 25–36. [DOI] [PubMed] [Google Scholar]
- 17.Guidotti, L. G., Borrow, P., Hobbs, M. V., Matzke, B., Gresser, I., Oldstone, M. B. & Chisari, F. V. (1996) Proc. Natl. Acad. Sci. USA 93, 4589–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wieland, S. F., Guidotti, L. G. & Chisari, F. V. (2000) J. Virol. 74, 4165–4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cavanaugh, V. J., Guidotti, L. G. & Chisari, F. V. (1997) J. Virol. 71, 3236–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cavanaugh, V. J., Guidotti, L. G. & Chisari, F. V. (1998) J. Virol. 72, 2630–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mason, W. & Litwin, S. (2002) in HBV Human Virus Guide, eds. Locarnini, S. & Lai, C. L. (Intl. Med. Press, London), pp. 99–113.
- 22.Carp, N. Z., Saputelli, J., Halbherr, T. C., Mason, W. S. & Jilbert, A. R. (1991) Lab. Anim. Sci. 41, 474–475. [PubMed] [Google Scholar]
- 23.Southern, E. M. (1975) J. Mol. Biol. 98, 503–517. [DOI] [PubMed] [Google Scholar]
- 24.Wahl, G. M., Stern, M. & Stark, G. R. (1979) Proc. Natl. Acad. Sci. USA 76, 3683–3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hornbuckle, W. E., Graham, E. S., Roth, L., Baldwin, B. H., Wickenden, C. & Tennant, B. C. (1985) Lab. Anim. Sci 35, 376–381. [PubMed] [Google Scholar]
- 26.Mason, W. S., Cullen, J., Moraleda, G., Saputelli, J., Aldrich, C. E., Miller, D. S., Tennant, B., Frick, L., Averett, D., Condreay, L. D. & Jilbert, A. R. (1998) Virology 245, 18–32. [DOI] [PubMed] [Google Scholar]
- 27.Jilbert, A. R. (2000) Methods Mol. Biol. 123, 177–193. [DOI] [PubMed] [Google Scholar]
- 28.Summers, J., Smith, P. M. & Horwich, A. L. (1990) J. Virol. 64, 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galibert, F., Chen, T. N. & Mandart, E. (1982) J. Virol. 41, 51–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, W. & Summers, J. (1999) J. Virol. 73, 9710–9717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, W. & Summers, J. (1998) J. Virol. 72, 8710–8717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang, W. & Summers, J. (1995) J. Virol. 69, 4029–4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang, W., Mason, W. S. & Summers, J. (1996) J. Virol. 70, 4567–4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Staprans, S., Loeb, D. D. & Ganem, D. (1991) J. Virol. 65, 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guidotti, L. G. & Chisari, F. V. (1999) Curr. Opin. Microbiol. 2, 388–391. [DOI] [PubMed] [Google Scholar]
- 36.Foster, W. K., Miller, D. S., Marion, P. L., Colonno, R. J., Kotlarski, I. & Jilbert, A. R. (2003) Antimicrob. Agents Chemother. 47, 2624–2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu, Y., Yamamoto, T., Cullen, J., Saputelli, J., Aldrich, C. E., Miller, D. S., Litwin, S., Furman, P. A., Jilbert, A. R. & Mason, W. S. (2001) J. Virol. 75, 311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Genovesi, E. V., Lamb, L., Medina, I., Taylor, D., Seifer, M., Innaimo, S., Colonno, R. J., Standring, D. N. & Clark, J. M. (1998) Antimicrob. Agents Chemother. 42, 3209–3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peek, S. F., Cote, P. J., Jacob, J. R., Toshkov, I. A., Hornbuckle, W. E., Baldwin, B. H., Wells, F. V., Chu, C. K., Gerin, J. L., Tennant, B. C. & Korba, B. E. (2001) Hepatology 33, 254–266. [DOI] [PubMed] [Google Scholar]
- 40.Luscombe, C., Pedersen, J., Uren, E. & Locarnini, S. (1996) Hepatology 24, 766–773. [DOI] [PubMed] [Google Scholar]
- 41.Fourel, I., Cullen, J. M., Saputelli, J., Aldrich, C. E., Schaffer, P., Averett, D. R., Pugh, J. & Mason, W. S. (1994) J. Virol. 68, 8321–8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.