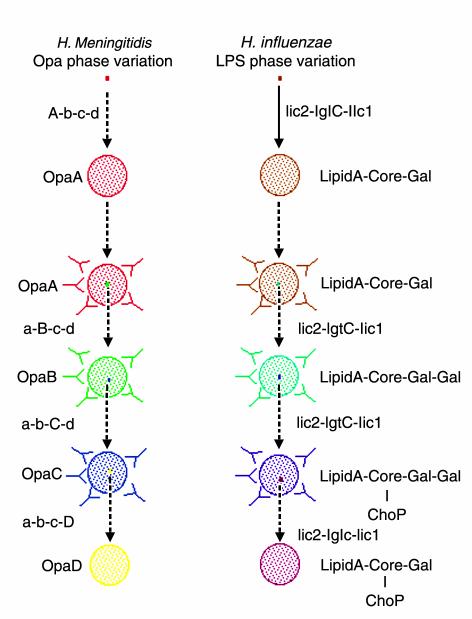
Figure 1.
Generation of antigenic variation by simple sequence contingency loci. Simple sequence contingency loci can rapidly generate variants during infections started by small numbers of bacteria that are essentially clonal. The events shown in (a) and (b) depict the hypothetical progression of two surface features that are targeted by the host immune system. In each, a single bacterium undergoes between 5 and 25 generations (solid line) to form a population within which there are a number of variants (27). After an indeterminate time (dotted lines), the host raises an antibody response against one determinant (a) or one part of a determinant (b) of these bacteria. A variant with an antigenically different determinant then escapes this immune response and replicates to form a new population containing variants that can then escape a second immune response. This process is repeated multiple times. (a) The opa genes of N. meningitidis provide an example of switching between determinants of similar function. Multiple, antigenically distinct forms of bacterial surface adhesins may be expressed in turn during the course of an infection, allowing the bacteria to evade the host’s immune defenses. (b) The LPS biosynthesis genes provide an example of switching between different modifications of a single determinant. The genotypes of the founder cell and the variants are indicated alongside the lines; genes that are switched on at a given point are shown in capital letters. The phenotype of the majority of cells in a population is indicated alongside the circles. In a, the product is a protein (note that the designations A to D for the opa genes [and proteins] are arbitrary and do not indicate particular opa genes); in b, the loci produce modifications to a basal LPS structure (16).