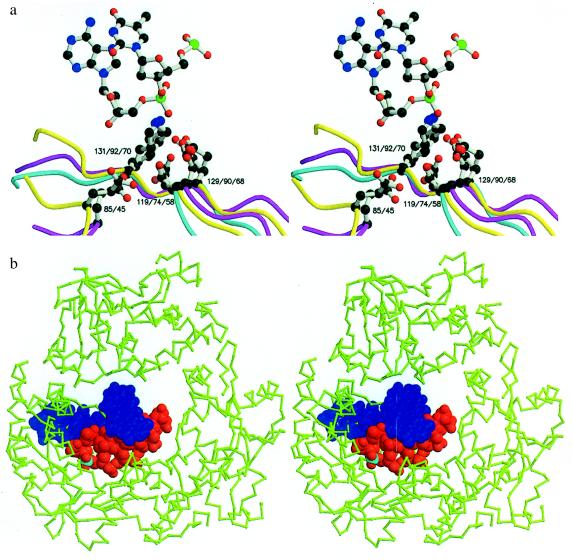
Figure 4.
(A) Stereoview showing the superposition of the active site residues of λ-exonuclease (yellow), (Glu-85, Asp-119, Glu-129, and Lys-131), PvuII (cyan) (Asp-58, Glu-68, and Lys-70), and EcoRV (magenta) (Glu-45, Asp-74, Asp-90, and Lys-92). The figure includes the central nucleotide (TA) from the co-crystal structure of EcoRV bound to its cognate DNA sequence (17). (B) Superposition on the trimeric structure of λ-exonuclease of the cognate DNA decamer from the EcoRV:DNA complex (17). To make the DNA resemble the substrate of λ-exonucleases, five nucleotides were removed from the 5′ end of the strand that is hydrolyzed. The next phosphate that would be attacked is shown in cyan. The DNA was aligned based on the correspondence between the active site residues of EcoRV and λ-exonuclease (A). In the stereoview, the wider end of the central channel through the λ-exonuclease trimer, i.e., the end of the channel that is thought to accept the double-strand DNA substrate, is toward the viewer. Figure created by using molscript (26) and rendered with raster3d (27, 28).