Abstract
Hairpin structures can inhibit DNA replication and are intermediates in certain recombination reactions. We have shown that the purified SbcCD protein of Escherichia coli cleaves a DNA hairpin. This cleavage does not require the presence of a free (3′ or 5′) DNA end and generates products with 3′-hydroxyl and 5′-phosphate termini. Electron microscopy of SbcCD has revealed the “head-rod-tail” structure predicted for the SMC (structural maintenance of chromosomes) family of proteins, of which SbcC is a member. This work provides evidence consistent with the proposal that SbcCD cleaves hairpin structures that halt the progress of the replication fork, allowing homologous recombination to restore DNA replication.
Keywords: hairpin nuclease/palindromic DNA/Rad50-Mre11
Long DNA palindromes inhibit DNA replication and are unstable in the genomes of Escherichia coli (see ref. 1), Bacillus (2), Streptococcus (3), Streptomyces (4), Saccharomyces cerevisiae (5–8), mice (9, 10), and humans (see ref. 11). These effects result from the formation of hairpin or cruciform structures by intrastrand base pairing (1). In E. coli, the inhibition of DNA replication is overcome significantly in sbcC or sbcD mutants (12, 13).
The sbcC and sbcD genes have been cloned and sequenced (14). SbcC has the characteristic molecular organization predicted for a polypeptide belonging to the SMC (structural maintenance of chromosomes) family (15, 16). This family of polypeptides is implicated in chromosome condensation and segregation (17–21), transcriptional repression (22), and genetic recombination (15, 23, 24). Members possess “Walker-A” and “Walker-B” nucleotide binding motifs (25) separated by an α-helical region (several hundred amino acids long) predicted to form two coiled-coil domains interrupted by a short spacer region (reviewed in refs. 16, 19, and 26). SbcC belongs to the subclass of SMC proteins implicated in genetic recombination that show more extensive sequence similarity in and around their ATP binding motifs and possess a conserved CXXC motif within their putative coiled-coil spacer region (15). This subclass includes the Rad50 polypeptide of S. cerevisiae (scRad50) (23), its human (27) and mouse (28) homologues (hRad50 and mRad50), and the product of bacteriophage T4 gene 46 (T4 gp46) (29). All of these polypeptides are involved in generating and/or processing DNA double-strand breaks. In vivo, scRad50 and hRad50 interact with the (sc or h) Mre11 polypeptide that also is involved in double-strand break repair (27, 30). The murine MRE11 gene has been shown to be essential for normal cell proliferation in embryonic stem cells (31). T4 gp46 has been shown genetically to interact with the product of gene 47 (gp47) (32). Both genes 46 and 47 are essential for recombination and DNA replication in T4 (33). ScMre11, hMre11 (34), and T4 gp47 are all putative nucleases that share sequence similarity with SbcD (15). They belong to a family of phosphoesterases, including various serine/threonine phosphatases, that contain the conserved sequence DXH(X25)GDXXD(X25)GNHD/E. This coupling of an SMC-family polypeptide to a phosphoesterase seems to be a common feature of these proteins involved in recombination and double-strand break repair.
SbcCD protein forms a large (1.2-MDa) complex that functions as an ATP-dependent double-strand DNA exonuclease and an ATP-independent single-strand endonuclease (35, 36). Density transfer and de-methylation studies have indicated that DNA replication is required before SbcCD can recognize DNA palindromes (37, 38). These observations have led to the proposal that SbcCD collapses replication forks by attacking hairpin structures that arise on the lagging-strand template and that broken replication forks are repaired by homologous recombination (1, 35). This proposal has been tested genetically by observing that a 240-bp palindrome introduced into the chromosome of E. coli causes cell death in sbcCD+ cells that are either recA, recB, or recC mutants (39).
Here, we test the prediction that SbcCD might recognize and cleave hairpin DNA. We demonstrate that SbcCD protein cleaves a hairpin DNA substrate at the 5′ side of the loop to yield products with 5′ phosphate and 3′ hydroxyl ends. This reaction does not require DNA termini. In addition, we provide physical evidence that SbcC(D) forms the head-rod-tail structure expected of an SMC protein.
MATERIALS AND METHODS
DNA Substrates.
To remove short chain termination products, all oligonucleotides (synthesized by Oswel DNA Service, University of Southampton) were run on 10% denaturing polyacrylamide gels and were visualized by UV shadowing (according to standard protocols; see ref. 40). DNA was recovered from excised bands by eluting at 60°C for 16 h in TE (10 mM Tris⋅HCl, pH 7.5/1 mM EDTA, pH 8.0) buffer, followed by ethanol precipitation. After 32P-end-labeling, oligonucleotides once again were gel purified and recovered as described above.
Hairpin substrate.
5′HP78 (5′-GTTTCTATTCAGCCCTTTGACGTAATCCAGCCCCGGGTTTTCCCGGGGCTGGATTACGTCAAAGGGCTGAATAGAAAC-3′) is a 78-nt oligomer capable of forming the hairpin structure, with a 37-bp stem and 4-nt loop, shown in Fig. 2. 3′HP78 is a 77-nt version of 5′HP78 that lacks the 3′-terminal cytosine nucleotide and contains a 5′-terminal phosphate group. After 3′ end-labeling, 3′HP78 possesses exactly the same sequence as 5′HP78 (see Fig. 2B). 5′HP78 was labeled (according to manufacturers’ instructions) at the 5′ end with [γ-32P]ATP (Amersham, 3,000 Ci/mmol) by using T4 polynucleotide kinase (Boehringer Mannheim) to yield a substrate with a specific activity of ≈6.7 × 106 cpm/μg total nucleic acid. 3′ end-labeled substrate was generated by filling in the 1-nt gap of 3′HP78 with [α-32P]dCTP (Amersham, 3,000 Ci/mmol) by using the Klenow fragment of DNA polymerase I (New England Biolabs) to yield a substrate with a specific activity of ≈7.4 × 106 cpm/μg total nucleic acid. The hairpin-forming oligonucleotides used here are based on those described by Beyert et al. (41). Restriction digestion and native PAGE were used to confirm the presence of hairpin secondary structure (data not shown).
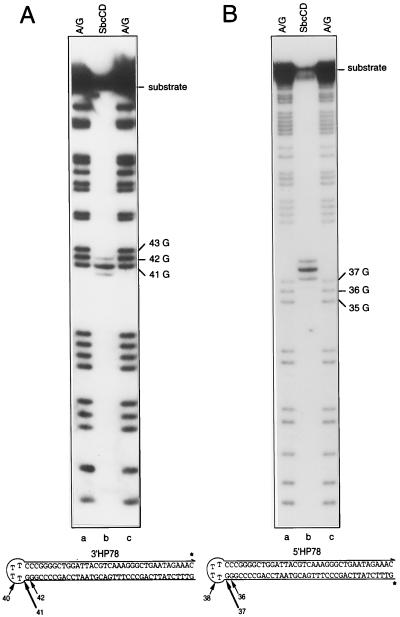
Figure 2.
5′- and 3′-labeled hairpin substrates incubated with SbcCD in the presence of ATPγS. ∗, the position of the 32P-label. The SbcCD product is flanked by A/G sequencing ladders. Fragments arising as a result of chemical cleavage at the three guanosines adjacent to the hairpin loop are indicated. (A) 3′-end-labeled hairpin (3′HP78) was incubated with SbcCD in the presence of ATPγS, at 16°C for 30 min, and reaction products were resolved on a polyacrylamide gel (lane b). A/G sequencing ladders of 3′HP78 are shown in lanes a and c. (B) 5′-end-labeled hairpin (5′HP78) was incubated with SbcCD in the presence of ATPγS, at 16°C for 30 min, and reaction products were resolved on a polyacrylamide gel (lane b). ∗, the position of the 32P-label. A/G sequencing ladders of 5′HP78 are shown in lanes a and c. A one-and-one-half base allowance was made to compensate for the nucleoside eliminated in the sequencing reaction.
Covalently closed dumbbell substrate.
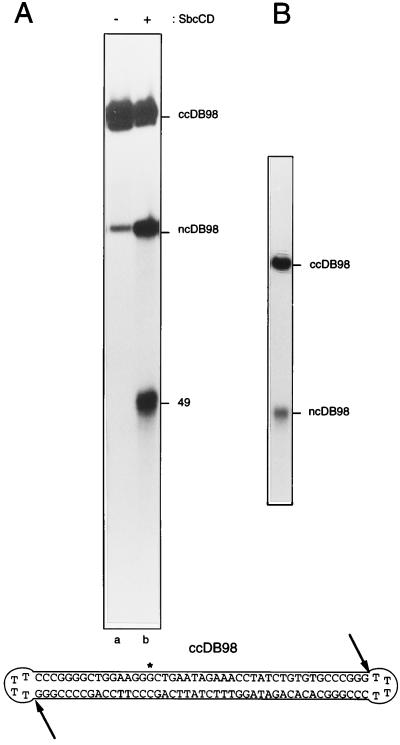
ncDB98 (5′-GCTGAATAGAAACCTATCTGTGTGCCCGGGTTTTCCCGGGCACACAGATAGGTTTCTATTCAGCCCTTCCAGCCCCGGGTTTTCCCGGGGCTGGAAGG-3′) is a 98-nt oligomer capable of forming a nicked-circular dumbbell (ncDB98). It was labeled (according to manufacturers’ instructions) at the 5′ end with [γ-32P]ATP by using T4 polynucleotide kinase to yield an oligonucleotide with a specific activity of ≈1.5 × 107 cpm/μg total nucleic acid. Covalently closed DB98 (ccDB98; see Fig. 4) was obtained by treating 2 pmol of boiled and snap-cooled 32P-labeled nicked-DB98 (ncDB98) with 400 units of T4 DNA ligase (New England Biolabs) at 37°C for 1 h in a 100-μl reaction volume. ccDB98 was separated from ncDB98 by denaturing gel electrophoresis. When ccDB98 is totally denatured it behaves as a single band (see Fig. 4B), with the slowly migrating characteristic of an open circle. Restriction digestion, chemical sequencing, and native PAGE were used to confirm that ccDB98 exists as a monomer–dumbbell (data not shown).
Figure 4.
SbcCD cleaves a covalently closed dumbbell substrate. (A) Internally labeled, covalently closed dumbbell substrate (ccDB98) was incubated without (lane a) or with (lane b) SbcCD in the presence of ATPγS, at 16°C for 30 min. ∗, the position of the 32P-label. Reaction products then were resolved on a denaturing polyacrylamide gel. The 49- and 98-nt fragments produced are indicated (49 and ncDB98). (B) Preparative 10% polyacrylamide gel showing the single product obtained after the ligation of ncDB98. The upper band is covalently closed dumbbell (ccDB98), the lower band is nicked-circular dumbbell (ncDB98), which failed to ligate.
Analysis of Hairpin Cleavage Termini.
All reactions were performed according to manufacturers’ recommendations.
5′-termini.
3′HP78 (3′-32P-labeled) DNA (≈1.5 nM) and SbcCD (2.5 nM) were incubated in SbcCD assay buffer (160 μl) containing 1 mM ATPγS for 30 min at 16°C. The reaction was stopped by the addition of EDTA to 10 mM, and DNA was recovered by ethanol precipitation. After resuspending in 10 μl of TE buffer, the DNA products were denatured by heating to 100°C for 3 min and were split into two 5-μl aliquots. The aliquots were added to two separate tubes containing alkaline phosphatase buffer (total volume 20 μl). After incubating for 60 min at 37°C, in the presence or absence of 4 units of shrimp alkaline phosphatase (United States Biochemical), the 32P-labeled products were analyzed by denaturing electrophoresis.
3′ termini.
5′HP78 (5′-32P-labeled) DNA (≈1.5 nM) and SbcCD (2.5 nM) were incubated in SbcCD assay buffer (80 μl) containing 1 mM ATPγS for 30 min at 16°C. The reaction was stopped by the addition of EDTA to 10 mM, and DNA was recovered by using a QIAquick nucleotide removal kit (Qiagen, Chatsworth, CA). After resuspending in 10 μl of 10 mM Tris⋅HCl (pH 7.5), the DNA products were denatured by heating to 100°C for 3 min and were split into two 5-μl aliquots. The aliquots were added to two separate tubes containing terminal transferase solution 1 (Boehringer Mannheim), 2.5 mM CoCl2, and 0.5 μM ddATP. After a 15-min incubation at 37°C in the presence or absence of 25 units of terminal transferase (Boehringer Mannheim), 32P-labeled products were analyzed by denaturing electrophoresis.
DNA Size Standards.
SbcCD cleavage sites were confirmed by comparing SbcCD products with an A/G sequencing ladder, produced by the chemical degradation of 3′-labeled 3′HP78 or 5′-labeled 5′HP78 (42). Chemical cleavage sites were confirmed by using an 8–32 base oligonucleotide sizing marker (Pharmacia) and DNA molecular weight marker V (Boehringer Mannheim).
Gel Electrophoresis.
Reactions were stopped by adding an equal volume of DNA loading buffer (1 mM EDTA, pH 8.0/95% formamide/0.01% bromophenol blue). Samples were heated to 100°C for 4 min before electrophoresis and were loaded as quickly as possible thereafter. To ensure conditions were as denaturing as possible, 10% polyacrylamide gels (0.4 mm thick) containing 7 M urea and 10% formamide were run on a Sequi-Gen Nucleic Acid Sequencing Cell (Bio-Rad) at ≈60°C for 1.5 h at 60 W using a TBE (89 mM Tris borate, pH 8.3/2 mM EDTA, pH 8.0) buffer system. DNA was visualized by autoradiography.
SbcCD Protein and Nuclease Assay.
SbcCD protein was prepared as described (36). Protein concentrations were estimated by using a protein assay kit (Bio-Rad) with BSA as a standard. The molar concentration of SbcCD was calculated assuming a stoichiometry of SbcC6:SbcD12. In the SbcCD nuclease assay, 2.5 nM protein and DNA (≈1.5 nM) were incubated for 30 min at the temperature indicated in a 20-μl reaction volume. The reactions were carried out in the presence of: 5 mM Mn2+, 1.25 mM DTT, 2% glycerol, 100 μg/ml BSA, 25 mM Tris (pH 7.5), and 1 mM ATPγS (or 1 mM ATP). Reactions were stopped and analyzed on a 10% denaturing polyacrylamide gel before drying and autoradiography.
Electron Microscopy.
Low angle rotary shadowing and unidirectional shadowing were performed as a service at the National Centre for Macromolecular Hydrodynamics, University of Leicester.
RESULTS
SbcCD Recognizes and Degrades a Hairpin Substrate.
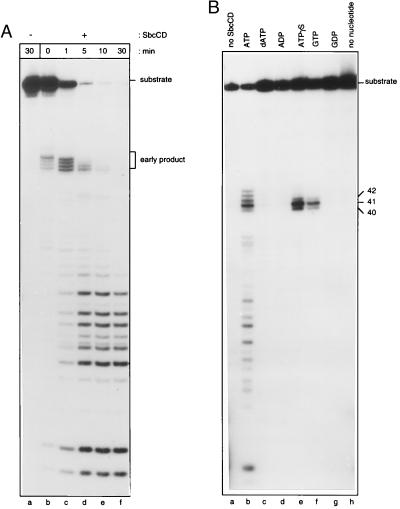
To test whether SbcCD was active on a hairpin substrate, a 3′-32P-end-labeled oligonucleotide (3′HP78) capable of forming a DNA hairpin with a 37-bp stem and a 4-nt loop (illustrated in Fig. 2A) was incubated with protein over a varying time period. After short incubation times (1 min or less) at 37°C, four main early cleavage products, ≈41–44 nt in length, were seen (Fig. 1A, lanes b and c). Longer incubation times (5–30 min) resulted in the disappearance of these fragments and the appearance of products ranging in size from ≈9–26 nt (Fig. 1A, lanes d–f). This experiment indicates that SbcCD nuclease is active on a hairpin substrate and suggests that initial cleavage occurs 41–44 nt distal to the 3′ end of the hairpin, in the vicinity of the stem-loop junction. It appears that SbcCD possesses a hairpin cleaving activity and that initial hairpin-cleavage products subsequently are degraded by the processive ATP-dependent double-strand exonuclease activity of the protein.
Figure 1.
Trapping the initial hairpin cleavage products generated by SbcCD. (A) An autoradiograph illustrating the DNA fragments produced by SbcCD over a 30-min time period. SbcCD was incubated with 3′-end-labeled hairpin substrate (3′HP78), in the presence of ATP at 37°C, for 0 min (lane b), 1 min (lane c), 5 min (lane d), 10 min (lane e), or 30 min (lane f). 3′HP78 also was incubated for 30 min in the absence of SbcCD (lane a). Early products produced by SbcCD are indicated. (B) Effect of various nucleotides on the SbcCD hairpin-nuclease activity. SbcCD was incubated with 3′-end-labeled hairpin substrate (3′HP78), at 16°C for 30 min, in the presence of various nucleotides. Reactions were performed in the presence of ATP (lane b), dATP (lane c), ADP (lane d), ATPγS (lane e), GTP (lane f), GDP (lane g), or no nucleotide (lane h). 3′HP78 also was incubated with ATP in the absence of SbcCD (lane a). Products were visualized by gel electrophoresis. The sizes of the fragments produced in the presence of ATPγS (lane e) and GTP (lane f) are indicated.
Initial Cleavage Products Can Be Separated from Subsequent Products.
Conditions were sought that would reduce (or inhibit) the processive double-strand exonuclease activity of SbcCD but retain hairpin cleavage activity. Protein was incubated with 3′HP78 (3′-end-labeled) hairpin oligonucleotide at 16°C in the presence of various nucleotide cofactors. No SbcCD activity was observed in the absence of nucleotide (Fig. 1B, lane h) or in the presence of dATP, ADP, or GDP (Fig. 1B, lanes c, d, and g, respectively). In the presence of ATP, products ranged in size from ≈9–44 nt (Fig. 1B, lane b). In the presence of ATPγS (a nonhydrolyzable analogue of ATP) or GTP (Fig. 1B, lanes e and f, respectively) one major cleavage product of ≈41 nt and two minor products of ≈40 and 42 nt were observed. This result indicated that ATPγS or GTP could be used to uncouple hairpin cleavage from double-strand exonuclease activity. It is possible that, under these conditions, the protein may not be turning over.
Initial Cleavage Occurs at the 5′ Side of a Hairpin Loop.
One major and two minor products were generated when hairpin DNA was incubated with SbcCD protein in the presence of ATPγS (Fig. 1B, lane e). To map these SbcCD cleavage sites more precisely, protein was incubated with hairpin substrates labeled at either their 3′ or 5′ termini. Cleavage sites were confirmed by comparing the SbcCD products with A/G sequencing ladders produced by the chemical degradation of 3′HP78 or 5′HP78 (both substrates are illustrated in Fig. 2 A and B). When 3′HP78 was incubated with SbcCD, the major product observed was a 41-nt DNA fragment. Two minor products of 40 and 42 nt also were seen (Fig. 2A, lane b). This result indicates that SbcCD preferentially cleaves 3′HP78 immediately adjacent to the loop/double-strand junction. To confirm that this was so, SbcCD was incubated with 5′-end-labeled hairpin 5′HP78. With this substrate, cleavage at the same junction should yield a major product of 37 nt and two minor products of 36 and 38 nt. All of these products can be seen in Fig. 2B, lane b. The migration of the SbcCD cleavage products relative to the A/G sequencing ladders was consistent with the presence of 5′-phosphate and 3′-hydroxyl termini at the cleavage site. To confirm this, enzymatic tests were performed (see below).
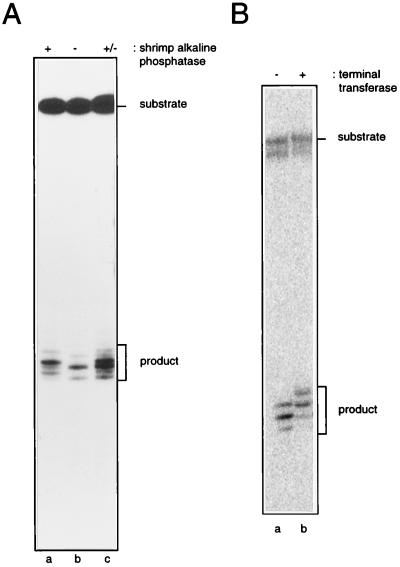
SbcCD Generates Products with 5′-Phosphate and 3′-Hydroxyl Termini.
To investigate the nature of the 5′-terminal group created by SbcCD, 3′-end-labeled hairpin (3′HP78) was incubated with SbcCD, and then the products were treated with shrimp alkaline phosphatase. After phosphatase treatment, all of the SbcCD products showed an altered mobility (Fig. 3A, lane a). This result indicates that SbcCD leaves a 5′-phosphate group when it cleaves DNA.
Figure 3.
Analysis of hairpin cleavage termini. (A) 5′ termini: 3′-end-labeled hairpin (3′HP78) was digested with SbcCD for 30 min at 16°C, in the presence of ATPγS, and the reaction products then were incubated with (lane a) or without (lane b) shrimp alkaline phosphatase. SbcCD cleavage products are indicated. A 1:1 mix of the samples loaded to lanes a and b were analyzed in lane c to demonstrate that the mobility shift observed was not a lane-specific artefact. (B) 3′-termini: 5′-end-labeled hairpin (5′HP78) was digested with SbcCD, in the presence of ATPγS, and the reaction products were incubated in the absence (lane a) or presence (lane b) of terminal transferase.
To determine the nature of the 3′-terminal group created by SbcCD, 5′-end-labeled hairpin (5′HP78) was incubated with SbcCD, and then the resulting products were treated with terminal transferase in the presence of ddATP. SbcCD products that had been treated with terminal transferase were seen to have increased in size by 1 nt (Fig. 3B, lane b), which demonstrates that SbcCD generates a 3′-hydroxyl group when it cleaves DNA. Therefore, SbcCD cleaves a hairpin substrate to leave products with 5′-phosphate and 3′-hydroxyl termini.
SbcCD Does Not Require a 3′ or 5′ Terminus for Activity.
To determine whether SbcCD was active on a molecule without free ends, protein was incubated with a covalently closed, internally labeled oligonucleotide (ccDB98; see Fig. 4). This substrate was capable of forming a dumbbell structure with a 45-bp duplex linked by two loop regions. ccDB98 was purified from a preparative gel as a single species (Fig. 4B). When ccDB98 was incubated at 16°C with ATPγS, in the presence of SbcCD, two major products of ≈98 and 49 nt (ncDB98) were observed (Fig. 4A, lane b). The size of both of these products strongly suggests that SbcCD is cleaving the covalently closed substrate at one or both junctions. In the absence of SbcCD, only a small amount of the band of ≈98 nt (ncDB98) was seen (Fig. 4A, lane a). This band is the expected product of radiolysis of ccDB98. A single nick anywhere in the backbone of this circular molecule would result in the appearance of a 98-nt fragment.
SbcC(D) Has a Head-Rod-Tail Structure.
SbcC is a polypeptide of 1,048 amino acids. Its sequence suggests that it contains ATP-binding motifs in globular regions at either end of the protein. These regions are separated by a 700-amino acid α-helical region likely to form a coiled-coil structure, broken by a conserved CXXC motif. This organization is typical of the head-rod-tail structure predicted for the SMC-family of proteins. Physical analysis revealed that SbcCD forms a 1.2-MDa complex (36). By using, EM we have demonstrated the existence of a structure consisting of two globular domains, linked by a long filamentous rod (Fig. 5). The globular domains at either end of the structure are ≈3.4 nm in diameter. The rod region between is ≈80 nm long and 1.5 nm wide (estimated by unidirectional shadowing). This region presumably comprises the amino acids of SbcC that are predicted to form an α-helical coiled-coil region. Additional experiments are required to determine the composition and stoichiometry of the structure visualized. Whether the D subunit remains associated to SbcC under the conditions of microscopy is unknown. For this reason, the D of SbcC(D) is indicated in parenthesis.
Figure 5.
Electron micrograph of SbcCD protein. SbcCD was visualized by using the low angle rotary shadowing method (Magnification, ×150,000.) Structures can be seen that consist of two globular domains linked by a long filamentous rod.
DISCUSSION
The SbcCD protein of E. coli has been shown to have exonuclease and endonuclease activities (35, 36), and genetic studies have suggested that it can act to generate double-strand breaks at DNA hairpins formed during DNA replication (39). These observations suggested that SbcCD might be able to recognize and cleave hairpin DNA. Here, we show that SbcCD does indeed cleave a hairpin substrate. Cleavage occurs on the 5′ side of the loop to yield products with 5′-phosphate and 3′-hydroxyl termini. In the presence of ATP, the initial products of cleavage are degraded by the processive double-strand exonuclease activity of SbcCD to yield smaller oligonucleotide products. In the presence of ATPγS or GTP, initial products accumulate, suggesting that nucleotide binding, but not hydrolysis, is required for hairpin cleavage.
Although SbcCD possesses an ATP-independent, single-strand endonuclease activity (35), a purine nucleoside triphosphate is required for hairpin cleavage. This requirement suggests that hairpin cleavage is not simply a consequence of the single-strand endonuclease activity of SbcCD. Hairpin cleavage is also distinct from the ATP-dependent double-strand exonuclease activity of SbcCD because no fragments released from the blunt (nonloop) end of the hairpin substrate were detected. SbcCD is active not only on a hairpin substrate but also on a closed circular dumbbell substrate. This indicates that SbcCD does not require 5′ or 3′ termini to cleave a molecule containing hairpin loops and suggests that it is some feature of the loop, or stem-loop junction, that is initially recognized. In contrast, the RecBCD enzyme of E. coli is an ATP-dependent, double-strand exonuclease and ATP-independent, single-strand endonuclease that is not active on dumbbell substrates (43). The ability of SbcCD to cleave a hairpin in the absence of a double-strand end is consistent with the recognition of a hairpin formed on the template strand of DNA replication. Cleavage of such a hairpin would lead to the formation of a double-strand break, resulting in collapse of the replication fork. The latter then could be reconstituted via a homologous recombination repair pathway.
DNA Replication and the Repair of Double-Strand Breaks.
Bacteriophage T4 and λ DNA replication can be initiated by homologous recombination at sites of DNA double-strand breaks (44–47), and it has been proposed that an important role of RecABCD-χ-mediated recombination in E. coli is the reconstitution of broken replication forks (48–50). Double-strand breaks may form during DNA replication because of the presence of a nick or gap in the template strand before its copying (50). Alternatively, replication arrest can lead to breakage of the fork (51–54), and palindromic sequences are known to cause replication arrest in vivo and in vitro (55, 56). It has been proposed that processing of hairpin secondary structures can result in the formation of double-strand breaks that then can be repaired by recombination (1, 35). For recombination to proceed, the loop at the apex of a hairpin must be removed because this structure can protect a DNA double-strand from recombination nucleases such as RecBCD enzyme (43). Homologous recombination indeed is required to repair the consequences of SbcCD attack of a 240-bp palindrome in the E. coli chromosome (39), and in S. cerevisiae, a 140-bp palindrome has been shown to act as a meiotic recombination hotspot by inducing a double-strand break (8).
Recombination Reactions That Require De-Protection of DNA Ends.
A hairpin DNA end is protected from attack by nucleases such as RecBCD that recognize 3′ or 5′ termini. Cleavage by SbcCD leads to homologous recombination (39), suggesting that the protein has a role in deprotecting a substrate and allowing it to enter the homologous recombination pathway. In S. cerevisiae, the Rad50/Mre11 complex has been suggested to play a different deprotecting role (39). In meiotic recombination, the SPO11 protein has been shown to become covalently attached to the 5′ ends of the initiating double-strand breaks (39). Resection of these ends requires Rad50/Mre11 (23). Perhaps the role of Rad50/Mre11 is to deprotect the double-strand break site by removing the bound SPO11 and to generate recombinogenic 3′ single-strand overhangs (57). Another possible example of an SbcCD family member acting to deprotect a covalently protected end has been described in bacteriophage T4. When mutants of T4 were sought that showed increased sensitivity to m-AMSA [4′-(9-acridinylamino)methanesulfon-m-anisidide], most mutations were found in genes that encode T4 recombination proteins, suggesting that recombination is involved in the repair of m-AMSA-induced covalent topoisomerase-DNA cleavage complexes (58, 59). Of the nine mutations identified, one was in gene 46 and another in a gene designated 47.1 (located upstream of gene 47). It was suggested that the mutation in gene 47.1 acts in a polar fashion on gene 47. Consistent with this explanation, a nearly complete deletion of gene 47.1 was constructed and was found not to cause hypersensitivity to m-AMSA (59).
In S. cerevisiae, RAD50 and MRE11 also play a role in nonhomologous end joining (60–62). Again, Rad50/Mre11 is acting at DNA ends although its role in end-joining is unknown. However, an interesting observation is that the same end-joining phenotype is obtained in hdf1 and ku80 mutants (61, 63). Hdf1 and scKu80 are the S. cerevisiae homologues of the mammalian Ku70 and Ku80 proteins that have been implicated in binding the hairpin DNA ends generated as intermediates in V(D)J recombination (64, 65). This raises the question of whether the human homologue of SbcCD (hRad50/hMre11) is involved in V(D)J recombination.
The factor that cleaves the hairpin intermediates in V(D)J recombination has not been identified. It has been established that an activity capable of nicking hairpin structures several nucleotides from the hairpin tip is present in murine lymphoid and nonlymphoid cells (10, 66). This is reminiscent of the preference SbcCD has for cleaving at the side of a hairpin loop. The presence of such an activity in nonlymphoid cells suggests that the ability to cleave a hairpin is required in a wider context than V(D)J recombination (10, 66). Whether or not hRad50/hMre11 is implicated directly in hairpin cleavage, the activity of SbcCD closely resembles the reaction predicted to be required. It will be interesting to compare the action of the mammalian hairpin cleaving nuclease(s) with SbcCD when they have been identified and characterized.
SbcC(D) Has the Head-Rod-Tail Structure Predicted for the SMC Family of Proteins.
EM has revealed that SbcC(D) has a filamentous structure with globular domains, of similar size, at either end of a rod. This tripartite head-rod-tail organization is typical of that predicted for the SMC-family of proteins and has been observed previously for two proteins implicated in chromosome segregation: the MukB protein of E. coli (67) and the BimC-related bipolar kinesin from Drosophila melanogaster (68). Neither of these proteins has been classified as belonging to the SMC family. In many SMC proteins, the potential coiled-coil domain is believed to be interrupted by a hinged spacer region. The putative coiled-coil of SbcC is broken by a conserved CXXC motif; yet, it is not obvious (from Fig. 5) that this sequence forms a hinged spacer within SbcC.
Despite their ubiquity, little is known of the biochemical activities of many SMC proteins. SbcC is an SMC protein that interacts with a non-SMC protein, SbcD (35, 36). Recently, several other SMC proteins also have been shown to exist in protein complexes with non-SMC proteins. The DPY27 polypeptide of Caenorhabditis elegans forms a dosage compensation complex with DPY26 and DPY28 (69, 70). The XCAP-C and XCAP-E polypeptides of Xenopus laevis form a chromosome condensation complex (condensin) with three other polypeptides (71). This complex exhibits a high affinity for structured DNA, such as cruciform DNA, and introduces positive supercoils into closed circular DNA in the presence of topoisomerase I (72). Two other SMC proteins, BSMC1 and BSMC2, exhibit DNA reannealing activity and form a recombination complex (RC-1) consisting of at least five polypeptides proposed to repair double-strand breaks in mammalian cells (24). This complex also recognizes and cleaves branched DNA substrates (73).
We have shown that SbcCD recognizes and cleaves a DNA hairpin substrate. However, the unusual protein structure would appear to be more complex than that required for such a relatively simple reaction. The SMC-family of proteins clearly are involved in many aspects of DNA metabolism in many different organisms. It is intriguing that several of these large SMC protein complexes, like SbcCD, recognize features of DNA secondary structure. Perhaps their common role is in targeting non-SMC proteins to these regions, where they perform varied roles important for maintaining genomic integrity.
Acknowledgments
We thank Bill Earnshaw and Dave Sherratt for critical reading of the manuscript; Karin Birkenkamp, Börries Kemper, and Petra Pfeiffer for help in designing and handling hairpin oligonucleotides; Stefan Hyman and Arthur Rowe for EM. This research has been supported by a project grant from the Biotechnology and Biological Sciences Research Council. L.A.K. is supported by a Biotechnology and Biological Sciences Research Council studentship.
ABBREVIATION
- SMC
structural maintenance of chromosomes
References
- 1.Leach D R F. BioEssays. 1994;16:893–900. doi: 10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 2.Peeters B P H, De Boer J H, Bron S, Venema G. Mol Gen Genet. 1988;212:450–458. doi: 10.1007/BF00330849. [DOI] [PubMed] [Google Scholar]
- 3.Behnke D, Malke H, Hartmann M, Walter F. Plasmid. 1979;2:605–616. doi: 10.1016/0147-619x(79)90058-1. [DOI] [PubMed] [Google Scholar]
- 4.Kieser T, Melton R. Gene. 1988;65:83–91. doi: 10.1016/0378-1119(88)90419-2. [DOI] [PubMed] [Google Scholar]
- 5.Henderson S T, Petes T D. Genetics. 1993;113:57–62. doi: 10.1093/genetics/134.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordenin D A, Lobachev K S, Degtyareva N P, Malkova A L, Perkins E, Resnick M A. Mol Cell Biol. 1993;13:5315–5322. doi: 10.1128/mcb.13.9.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruskin B, Fink G R. Genetics. 1993;133:43–56. doi: 10.1093/genetics/134.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nag D K, Kurst A. Genetics. 1997;146:835–847. doi: 10.1093/genetics/146.3.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collick A, Drew J, Penberth J, Bois P, Luckett J, Scaerou F, Jeffreys A, Reik W. EMBO J. 1996;15:1163–1171. [PMC free article] [PubMed] [Google Scholar]
- 10.Akgun E, Zahn J, Baumes S, Brown G, Liang F, Romanienko P J, Lewis S, Jasin M. Mol Cell Biol. 1997;17:5559–5570. doi: 10.1128/mcb.17.9.5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kramer P R, Stringer J R, Sinden R R. Nucleic Acids Res. 1996;24:4234–4241. doi: 10.1093/nar/24.21.4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chalker A F, Leach D R F, Lloyd R G. Gene. 1988;71:201–205. doi: 10.1016/0378-1119(88)90092-3. [DOI] [PubMed] [Google Scholar]
- 13.Gibson F P, Leach D R F, Lloyd R G. J Bacteriol. 1992;174:1222–1228. doi: 10.1128/jb.174.4.1222-1228.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naom I S, Morton S J, Leach D R F, Lloyd R G. Nucleic Acids Res. 1989;17:8033–8045. doi: 10.1093/nar/17.20.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharples G J, Leach D R F. Mol Microbiol. 1995;17:1215–1220. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirano T, Mitchison T J, Swedlow J R. Curr Opin Cell Biol. 1995;7:329–336. doi: 10.1016/0955-0674(95)80087-5. [DOI] [PubMed] [Google Scholar]
- 17.Strunikov A V, Larinov V, Koshland D. J Cell Biol. 1993;123:1635–1648. doi: 10.1083/jcb.123.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hirano T, Mitchison T J. Cell. 1994;79:449–458. doi: 10.1016/0092-8674(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 19.Saitoh N, Goldberg I G, Wood E R, Earnshaw W C. J Cell Biol. 1994;127:303–318. doi: 10.1083/jcb.127.2.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strunikov A V, Hogan E, Koshland D. Genes Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- 22.Chuang P T, Albertson D G, Meyer B J. Cell. 1994;79:459–474. doi: 10.1016/0092-8674(94)90255-0. [DOI] [PubMed] [Google Scholar]
- 23.Alani E, Subbiah S, Kleckner N. Genetics. 1989;122:47–57. doi: 10.1093/genetics/122.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessberger R, Riwar B, Baechtold H, Akhmedov A T. EMBO J. 1996;15:4061–4068. [PMC free article] [PubMed] [Google Scholar]
- 25.Walker J, Saraste M, Runswick M, Gay N. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gasser S M. Curr Biol. 1995;5:357–360. doi: 10.1016/s0960-9822(95)00071-6. [DOI] [PubMed] [Google Scholar]
- 27.Dolganov G M, Maser R S, Novikov A, Tosto L, Chong S, Bressan D A, Petrini J H J. Mol Cell Biol. 1996;16:4832–4841. doi: 10.1128/mcb.16.9.4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim K K, Daud A I, Wong S C, Pajak L, Tsai S C, Wang H, Henzel W J, Field L J. J Biol Chem. 1996;271:29255–29264. doi: 10.1074/jbc.271.46.29255. [DOI] [PubMed] [Google Scholar]
- 29.Leach D R F, Lloyd R G, Coulson A F C. Genetica. 1992;87:95–100. doi: 10.1007/BF00120998. [DOI] [PubMed] [Google Scholar]
- 30.Johzuka K, Ogawa H. Genetics. 1995;139:1521–1532. doi: 10.1093/genetics/139.4.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao Y, Weaver D T. Nucleic Acids Res. 1997;25:2985–2991. doi: 10.1093/nar/25.15.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mickelson C, Wiberg J S. J Virol. 1981;40:65–77. doi: 10.1128/jvi.40.1.65-77.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kreuzer K N, Saunders M, Weislo L J, Kreuzer H W E. J Bacteriol. 1995;177:6844–6853. doi: 10.1128/jb.177.23.6844-6853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Petrini H J, Walsh M E, DiMare C, Chen X-N, Kornberg J R, Weaver D T. Genomics. 1995;29:80–86. doi: 10.1006/geno.1995.1217. [DOI] [PubMed] [Google Scholar]
- 35.Connelly J C, Leach D R F. Genes to Cells. 1996;1:285–291. doi: 10.1046/j.1365-2443.1996.23024.x. [DOI] [PubMed] [Google Scholar]
- 36.Connelly J C, deLeau E S, Okely E A, Leach D R F. J Biol Chem. 1997;272:19819–19826. doi: 10.1074/jbc.272.32.19819. [DOI] [PubMed] [Google Scholar]
- 37.Shurvinton C E, Stahl M M, Stahl F W. Proc Natl Acad Sci USA. 1987;84:1624–1628. doi: 10.1073/pnas.84.6.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindsey J C, Leach D R F. J Mol Biol. 1989;206:7024–7027. doi: 10.1016/0022-2836(89)90584-6. [DOI] [PubMed] [Google Scholar]
- 39.Leach D R F, Okely E A, Pinder D J. Mol Microbiol. 1997;26:597–606. doi: 10.1046/j.1365-2958.1997.6071957.x. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Frisch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 41.Beyert N, Reichenberger S, Peters M, Hartung M, Gottlich B, Goedecke W, Vielmetter W, Pfeiffer P. Nucleic Acids Res. 1994;22:1643–1650. doi: 10.1093/nar/22.9.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Papavassiliou A G. In: Methods in Molecular Biology. Kneale G G, editor. Vol. 30. Clifton, NJ: Humana; 1994. pp. 43–78. [DOI] [PubMed] [Google Scholar]
- 43.Taylor A F, Smith G R. J Biol Chem. 1995;41:24459–24467. doi: 10.1074/jbc.270.41.24459. [DOI] [PubMed] [Google Scholar]
- 44.Luder A, Mosig G. Proc Natl Acad Sci USA. 1982;79:1101–1105. doi: 10.1073/pnas.79.4.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mosig G. In: Relationship of T4 DNA replication and recombination. In Bacteriophage T4. Mathews C, Kutter E, Mosig G, Berget P, editors. Washington DC: Am. Soc. Microbiol.; 1983. pp. 120–130. [Google Scholar]
- 46.Stahl F W, Kobayashi I, Stahl M. J Mol Biol. 1984;181:199–209. doi: 10.1016/0022-2836(85)90085-3. [DOI] [PubMed] [Google Scholar]
- 47.Stahl F W, Stahl M M. Genetics. 1986;113:1–12. doi: 10.1093/genetics/113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Asai T, Bates D B, Kogoma T. Cell. 1994;78:1051–1061. doi: 10.1016/0092-8674(94)90279-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuzminov A, Schabtach E, Stahl F W. EMBO J. 1994;13:2764–2776. doi: 10.1002/j.1460-2075.1994.tb06570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuzminov A. Mol Microbiol. 1995;16:373–384. doi: 10.1111/j.1365-2958.1995.tb02403.x. [DOI] [PubMed] [Google Scholar]
- 51.Bierne H, Ehrlich S D, Michel B. EMBO J. 1991;10:2699–2705. doi: 10.1002/j.1460-2075.1991.tb07814.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bierne H, Michel B. Mol Microbiol. 1994;13:17–23. doi: 10.1111/j.1365-2958.1994.tb00398.x. [DOI] [PubMed] [Google Scholar]
- 53.Kuzminov A. BioEssays. 1995;17:733–741. doi: 10.1002/bies.950170810. [DOI] [PubMed] [Google Scholar]
- 54.Michel B, Ehrlich S D, Uzest M. EMBO J. 1997;16:430–438. doi: 10.1093/emboj/16.2.430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.La Duca R J, Fay P J, Chuang C, McHenry C S, Banbara R A. Biochemistry. 1983;22:5177–5188. doi: 10.1021/bi00291a018. [DOI] [PubMed] [Google Scholar]
- 56.Weaver D T, DePamphlis M L. J Mol Biol. 1984;180:961–986. doi: 10.1016/0022-2836(84)90266-3. [DOI] [PubMed] [Google Scholar]
- 57.Keeney S, Giroux C N, Kleckner N. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 58.Neece S H, Carles-Kinch K, Tomso D J, Kreuzer K N. Mol Microbiol. 1996;20:1145–1154. doi: 10.1111/j.1365-2958.1996.tb02635.x. [DOI] [PubMed] [Google Scholar]
- 59.Woodworth D L, Kreuzer K N. Genetics. 1996;143:1081–1090. doi: 10.1093/genetics/143.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Moore J K, Haber J E. Mol Cell Biol. 1996;16:2164–2173. doi: 10.1128/mcb.16.5.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Milne G T, Jin S, Shannon K B, Weaver D T. Mol Cell Biol. 1996;16:4189–4198. doi: 10.1128/mcb.16.8.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tsukamoto Y, Kato J-I, Ikeda H. Genetics. 1996;142:383–391. doi: 10.1093/genetics/142.2.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tsukamoto Y, Kato J-I, Ikeda H. Nucleic Acids Res. 1996;24:2067–2072. doi: 10.1093/nar/24.11.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Taccioli G, Gottlieb M T, Blunt T, Priestly A, Demengeot J, Mizuta R, Lehman A R, Alt F W, Jackson S P, Jeggo P A. Science. 1994;266:1442–1445. doi: 10.1126/science.8073286. [DOI] [PubMed] [Google Scholar]
- 65.Boubnov N V, Hall K T, Wills Z, Lee S E, He D M, Benjamin D M, Pulaski C R, Band H, Reeves W, Hendrickson E A, et al. Proc Natl Acad Sci USA. 1995;92:890–894. doi: 10.1073/pnas.92.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis S M. Proc Natl Acad Sci USA. 1994;91:1332–1336. doi: 10.1073/pnas.91.4.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Niki H, Imamura R, Kitaoka M, Yamanaka K, Ogura T, Hiraga S. EMBO J. 1992;13:5101–5109. doi: 10.1002/j.1460-2075.1992.tb05617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kashina A S, Baskin R J, Cole D G, Wedaman K P, Saxton W M, Scholey J M. Nature (London) 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chuang P T, Lieb J D, Meyer B J. Science. 1996;274:1736–1739. doi: 10.1126/science.274.5293.1736. [DOI] [PubMed] [Google Scholar]
- 70.Lieb J, Capowski E, Meneely P, Meyer B. Science. 1996;274:1732–1736. doi: 10.1126/science.274.5293.1732. [DOI] [PubMed] [Google Scholar]
- 71.Hirano T, Kobayashi R, Hirano M. Cell. 1997;89:511–521. doi: 10.1016/s0092-8674(00)80233-0. [DOI] [PubMed] [Google Scholar]
- 72.Kimura K, Hirano T. Cell. 1997;90:625–634. doi: 10.1016/s0092-8674(00)80524-3. [DOI] [PubMed] [Google Scholar]
- 73.Jessberger R, Chui G, Linn S, Kemper B. Mutation Res. 1996;350:217–227. doi: 10.1016/0027-5107(95)00106-9. [DOI] [PubMed] [Google Scholar]