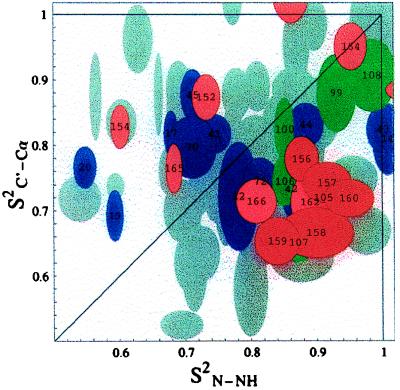
Figure 2.
The order parameters S2C′-Cα versus S2N-NH for E. coli flavodoxin. Data for most peptide planes are shown as overlapping gray ellipsoids representing the experimental uncertainties. The error ranges (± one SD) were obtained by computing the order parameters for a range of synthetic input data with distributions corresponding to the primary uncertainties of the raw input relaxation data as estimated from spectral signal-to-noise ratios. Red and pink ellipsoids correspond to the peptide planes of helix 5 as indicated by the residue numbers. Residues 163 and 153 are on the top and right-hand side of the figure, respectively. The green ellipsoids show helix 4, and the purple ellipsoids show all other helical residues for which data was obtained. Note: The coordinates of data point i correspond to the order parameters of the vectors C′(i)-Cα(i) and N(i + 1)-NH(i + 1). The order parameters S2N-NH were measured from 15N T1, T1ρ, and nuclear Overhauser effect data. The T1ρ data for these residues were found to be independent of spin locking rf power. Hence, we conclude that the order parameters for these residues have no exchange component, and therefore, that the anisotropic motions are confined to the nanosecond to picosecond time domain. The order parameters S2C′-Cα were measured from Cα-C′ nuclear Overhauser effect experiments combined with 13C′ T1 relaxation as described in ref. 16. Because T1ρ and T2 were not used, the C′ order parameters also have no exchange component. Experimental details are given in ref. 29.