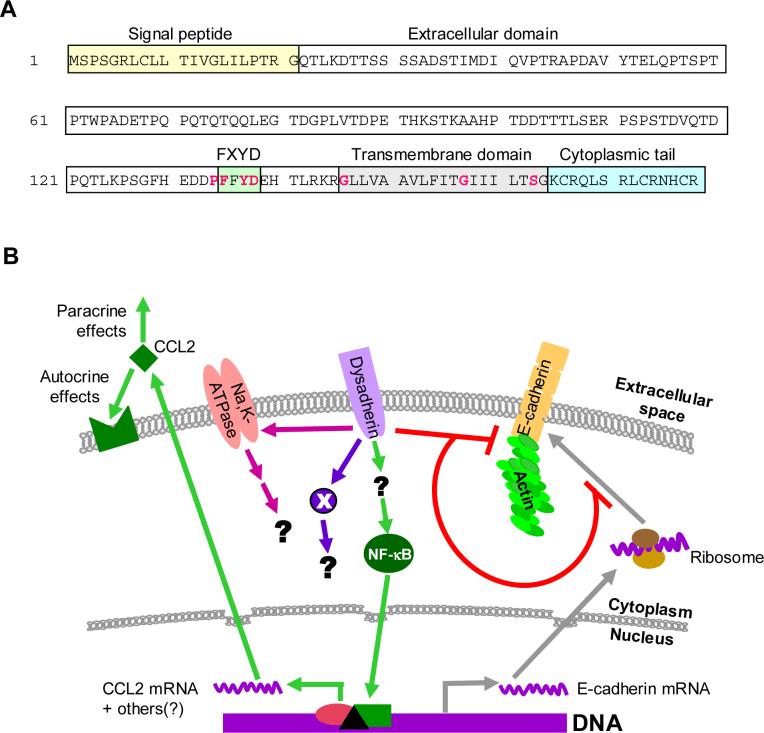
Fig. 1. Dysadherin structure, and a model for the role of dysadherin in cancer progression.
A. Primary sequence and domain structure for human dysadherin. Dysadherin is a single-pass transmembrane protein of 178 amino acids, with a long extracellular domain, a transmembrane domain and a short cytoplasmic tail. The highest conservation between FXYD family members lies in the region of the FXYD motif and the transmembrane domain. Residues that are absolutely conserved between all family members are shown in red. The extracellular domain is heavily and heterogeneously glycosylated, carrying up to 30KDa of O-linked carbohydrate in some tumor cells. B. Model for dysadherin action. Dysadherin predominantly resides in the plasma membrane. There is convincing evidence that dysadherin can reduce cell-cell adhesion by blocking expression of E-cadherin at a post-transcriptional level, and/or by down-regulating the association of E-cadherin with the actin cytoskeleton. Dysadherin can also affect invasion and metastasis by E-cadherin-independent mechanisms. One such mechanism involves upregulating expression of the chemokine CCL2, which then exerts autocrine and paracrine tumor-promoting effects in the tumor bed. The detailed molecular mechanisms that couple dysadherin expression to down-stream events that promote tumor progression are currently not known. However, dysadherin expression is associated with enhanced activity of the NFκB pathway in a breast cancer model system. In non-transformed cells, dysadherin interacts with and modulates the activity of the Na+,K+-ATPase. It remains to be determined whether this interaction occurs in tumor cells or what the functional consequences may be. Finally, it is likely that dysadherin also regulates the activity of additional downstream effector molecules (indicated by X) that have yet to be identified.