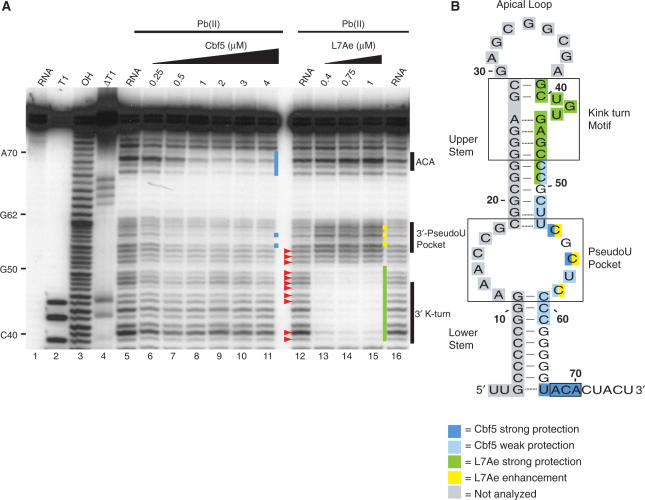
Figure 4.
Lead-induced cleavage footprinting of Pf9 RNA, and Cbf5–Pf9 and L7Ae–Pf9 sub-complexes. (A) 5′-end labelled Pf9 was incubated in the absence (lanes 5, 12, 16) or presence of increasing concentrations of Cbf5 (lanes 6–11) or L7Ae (lanes 13–15) and subjected to lead (II)-induced cleavage. Lane 1 is undigested RNA and lanes 3, 2 and 4 are size markers generated by alkaline hydrolysis (OH), and RNase T1 digestion under non-denaturing (T1) and denaturing conditions (ΔT1), respectively. Blue and green bars indicate regions of strong Cbf5 and L7Ae protection. Yellow bars indicate cleavage enhancements observed with L7Ae. Red arrowheads indicate unexpected cleavages in the upper stem of the guide RNA in the absence of protein. (B) Summary of cleavage protections and enhancements in the context of Pf9 RNA secondary structure model (as in Figure 1).