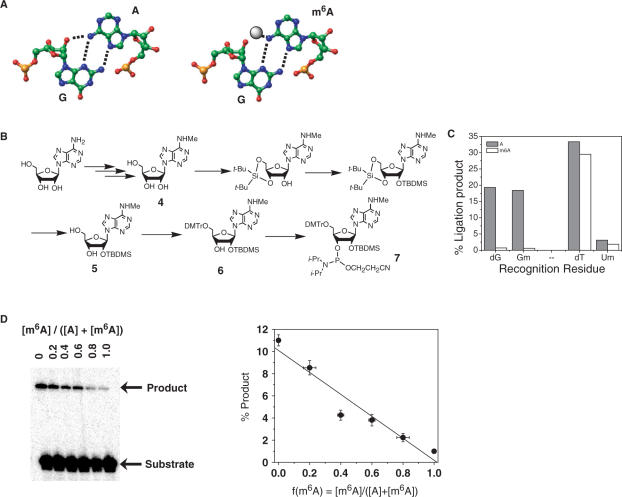
Figure 3.
Identifying a recognition residue for m6A. (A) The sheared G–A base pair found in many RNA structures (left). Within this purine–purine base pair N6H(A) and 2′OH(G) reside in close proximity to each other. The methyl group of m6A (right) might sterically clash with the phosphate backbone of G. (B) Synthetic scheme for N6-methyl-rA phosphoramidite. (C) Efficiency of A and m6A-directed ligation using different recognition residues. The 30-mer RNA templates contained at position 15 either unmodified adenosine (A) or N6-methyladenosine (m6A). Substrates contained one of the following recognition residues: dG (2′-deoxyguanosine), Gm (2′-methoxyguanosine), dT (thymidine) or Um (2′-methoxyuridine). (D) Ligation yield correlates linearly with the fraction of adenosine methylation f(m6A). Ligation reactions contained adenosine and N6-methyladenosine 30-mer RNA templates mixed together in defined ratios as indicated. Linear fit has a r-value of 0.964 and P-value of 0.002.