Abstract
The trans-activating responsive (TAR) RNA element located in the 5′ untranslated region of the HIV-1 genome is a 57-nt imperfect stem-loop essential for the viral replication. TAR regulates transcription by interacting with both viral and cellular proteins. RNA hairpin aptamers specific for TAR were previously identified by in vitro selection [Ducongé,F. and Toulmé,J.J. (1999) In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA, 5, 1605–1614]. These aptamers display a 5′-GUCCCAGA-3′ consensus apical loop, partially complementary to the TAR one, leading to the formation of a TAR–aptamer kissing complex. The conserved GA combination (underlined in the consensus sequence) has been shown to be crucial for the formation of a highly stable complex. To improve the nuclease resistance of the aptamer and to increase its affinity for TAR, locked nucleic acid (LNA) nucleotides were introduced in the aptamer apical loop. LNA are nucleic acids analogues that contain a 2′-O,4′-C methylene linkage and that raise the thermostablity of duplexes. We solved the NMR solution structure of the TAR–LNA-modified aptamer kissing complex. Structural analysis revealed the formation of a non-canonical G•A pair leading to increased stacking at the stem-loop junction. Our data also showed that the introduction of LNA residues provides an enhanced stability while maintaining a normal Watson–Crick base pairing with a loop–loop conformation close to an A-type.
INTRODUCTION
The trans-activating responsive (TAR) RNA element of HIV-1, located at the 5′ untranslated region of the viral genome, is a 57-nt imperfect stem-loop essential for HIV replication (1–4). TAR regulates transcription by interacting with viral and cellular proteins. The upper part of TAR constitutes the binding site of the viral protein Tat that recruits cellular proteins from the positive transcription elongation factor (P-TEFb) complex (5). The TAR element displays a highly conserved 6-nt loop 5′-CUGGGA-3′ (6) and constitutes a valid target for designing ligands that could inhibit TAR–protein interactions, thus preventing the development of the virus.
The systematic evolution of ligands by exponential enrichment (SELEX) approach was used against TAR to generate high affinity ligands. DNA and RNA aptamers specific for TAR were identified by in vitro selection (7,8). These aptamers, folded as stem-loop, displayed a 5′-ACTCCCAT-3′ (for DNA) or 5′-GUCCCAGA-3′ (for RNA) consensus sequence in the apical loop, partially complementary to the TAR one, leading to the formation of TAR–aptamer ‘kissing’ complexes (9) (Figure 1A). Similar motifs regulate gene expression in different organisms. Loop–loop interactions have been shown to regulate the copy number of the plasmid ColE1 (10). The dimerization and encapsidation of retroviral RNAs of the Moloney murine leukaemia virus are initiated through the formation of a RNA kissing complex (11). The dimerization of the HIV genome is also initiated at a conserved stem-loop structure that forms a kissing complex (12–17).
Figure 1.
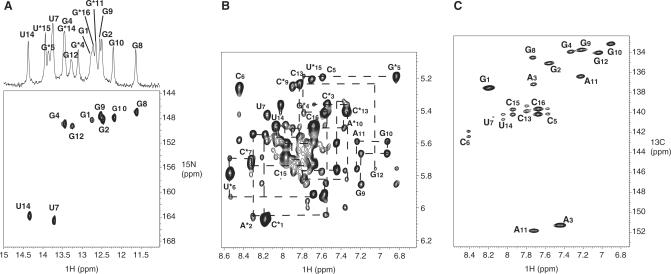
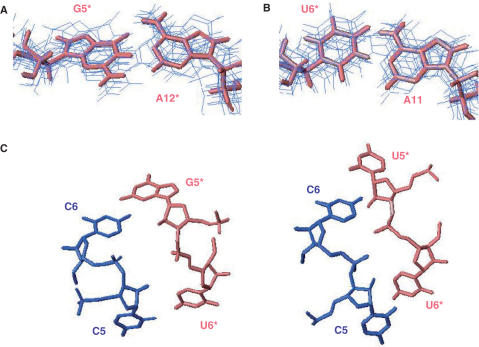
Comparison of TAR/R06(GA) and TAR/LR06(GA) complexes. (A) Secondary structure of TAR/R06(GA) (left) and TAR/LR06(GA) (right) complexes based on exchangeable and non-exchangeable proton NMR data. Two Gs were introduced at the 5′-end of R06(GA) to improve the yield of the transcription. LNAs residues are in red (asterisk indicates aptamer residues). (B) View of a C ribonucleotide residue (top) and of a C-LNA residue (bottom). (C) Imino proton region of 1D spectra of TAR/R06(GA) (top) and TAR/LR06(GA) (bottom).
The loop of the TAR RNA aptamer, termed R06(GA), is closed by selected conserved G and A residues on the 5′ and the 3′ sides, respectively. Mutations of these residues decreased the stability of the TAR–aptamer complex (18). To improve the nuclease resistance of the aptamer, chemically modified analogues of R06(GA) were investigated (19–21). Among them, the locked nucleic acid (LNA) has received particular attention (22–25). LNA residues are ribonucleotide analogues containing a methylene linkage between the 2′-O and 4′-C of the ribose ring that locks the sugar moiety in a C3′-endo-conformation (26) (Figure 1B). This generates the most stable hybrids ever characterized with a ΔTm of +3 and +10°C per LNA residue upon binding to DNA and RNA, respectively (27–29). Structural studies have pointed out interesting properties of oligonucleotides containing LNA and DNA residues. LNA changes the sugar conformation of adjacent DNA residues to the N-type making DNA/LNA hybrids good mimics of RNA (21,27–32). This effect reaches a maximum for oligonucleotides containing <50% of LNA residues. These properties of LNA were considered to generate an aptamer derived from the parent RNA aptamer R06(GA), with improved binding properties (21).
Only a limited number of LNA modifications in the aptamer loop are tolerated for generating a ligand recognizing the TAR target (21). In a recent work, all possible combinations (26) involving LNA and 2′-O-methyl residues in the anti-TAR aptamer loop (except closing G, A residues) were tested (33). Results pointed out that the affinity of these aptamers for TAR decreased with increasing LNA residues in the loop. Moreover, stability also depends on the location of LNA residues. Three aptamers with one or two LNAs at positions 5 or 5,6 or 5,7 (the G residue on the 5′ side of the loop being numbered 1) display affinities for TAR one or two orders of magnitude higher than the parent RNA aptamer. One of these combinations, containing two LNAs at positions 5 and 6 in the loop was able to inhibit the TAR-dependent expression of a luciferase reporter gene in cultured cells (33). On the other hand, no inhibitory effect was observed with an aptamer containing four LNAs in the loop at positions 2, 4, 6 and 7. Preliminary structural studies by nuclear magnetic resonance (NMR) revealed that the position of LNAs in the loop induced structural modifications at the stem-loop junctions (34).
To elucidate the origin of the stabilization due to the GA combination and to the introduction of LNAs in the loop, the solution structure of the kissing complex formed between TAR and the LNA/RNA chimeric aptamer, LR06(GA), with LNAs at positions 5 and 6 in the loop was solved by NMR. Structural analysis reveals that the stability results from the formation of a G•A pair that increased the stacking at the stem-loop junction. In addition, the data show that the LNA residues at positions 5 and 6 also provide additional stability compared to the parent RNA aptamer.
MATERIALS AND METHODS
Preparation of RNA samples for NMR
Unlabelled LNA/RNA chimeric aptamer LR06(GA) was purchased from Eurogentec (Belgium). Milligram quantities of TAR RNA were prepared unlabelled or 13C-15N labelled by in vitro transcription with T7 RNA polymerase from an oligonucleotide template containing a 2′-O-methyl modification at position 2 (35). Unlabelled R06(GA) was also prepared. Two G were introduced at the 5′-end to improve the yield of the transcription (Figure 1A). Labelled NTPs (nucleotide triphosphate) were purchased from Spectra Stable Isotopes (Columbia, USA). LR06(GA), R06(GA) and TAR were purified as described by Puglisi and Wyatt (36). After electroelution and ethanol precipitation, resuspended RNAs were then dialysed for 48 h against the buffer used for NMR experiments. Samples were concentrated by lyophilization and resuspended in 90%/10% H2O/D2O for experiments involving exchangeable protons and in 100% D2O for non-exchangeable protons experiments. Each sample was refolded by heating at 95°C (2 min) and snap-cooled at 4°C. Complexes were formed by addition of TAR to LR06(GA) monitoring the imino region of 1D spectra. At each point of the titration, the sample was heated at 95°C and snap-cooled at 4°C.
Proton and heteronuclear NMR
NMR experiments were recorded at 500 MHz on a Bruker Avance spectrometer equipped with a BBI (broad band inverse) z-gradient probe. NMR data were processed using TopSpin (Bruker) and were analysed using Sparky software packages (37). NMR experiments were performed in 10 mM sodium phosphate buffer (pH 6.4). The concentration of unlabelled and labelled RNAs samples ranged from 0.8 to 1 mM. Samples volumes were 280 μl in Shigemi NMR tubes.
1H, 13C and 15N assignments were obtained using standard homonuclear and heteronuclear methods. NMR data were acquired at 4 or 15°C for exchangeable protons and at 15°C for non-exchangeable protons experiments. Solvent suppression for samples in 90%/10% H2O/D2O was achieved using the WATERGATE and ‘Jump and Return’ sequences (38–40). The residual HDO resonance in D2O was suppressed using low power pre-saturation. Two-dimensional NOESY spectra in 90%/10% H2O/D2O were acquired with mixing time of 300, 150 and 50 ms. Base pairing was established via sequential nuclear Overhauser effects (NOEs) observed in 2D NOESY spectra at different mixing times. NOESY spectra with mixing times of 50, 150, 200 and 400 ms in D2O were acquired at 15°C.
Heteronuclear NMR spectra were measured at 15°C in 100% D2O at the exception of the 1H-15N heteronuclear single quantum coherence (HSQC) experiment that was acquired at 4°C in 90%/10% H2O/D2O (41). 13C or 15N decoupling during acquisition time was achieved using globally optimized alternating phase rectangular pulse (GARP) composite pulse sequence. The assignment of non-exchangeable proton was achieved using 2D NOESY experiment at different mixing times, 2D TOCSY, 2D COSY-DQF (double quantum-filtered correlated spectroscopy) and 3D NOESY-NOESY (42–44). The assignment of labelled TAR was completed using 1H-13C HSQC, 3D HCCH-TOCSY and 3D NOESY-HMQC (41,45,46). Resonance assignments are reported relative to TSP(3-(trimethylsilyl) propionic acid-d4 sodium salt) and to known chemical shifts.
Distance and dihedral restraints for structure calculation
Distance restraints involving non-exchangeable protons were derived from visual inspection of cross-peak intensities in 50, 150, 200 and 400 ms NOESY experiments using the H5/H6 cross-peak of pyrimidine as internal standard. Inter-proton distances derived from NOE cross-peak volumes were classified into four distance bound ranges: strong (1.8–3.0 Å), medium (3.0–4.5 Å), weak (4.0–5.5 Å) and very weak (4.5–7.0 Å). Hydrogen bonding restraints were used for Watson–Crick base pairs: from G1•C16 to C5•G12, C1*•G16* to G4*•C13* and C6•G11* to G10•C7*. One hydrogen bond was added between A11(N1) and U6*(H3).
Classical A-helix angles values were used for G1 to G4, C13 to C16, C1* to C3* and G14* to G16* (Figure 1A): α = −65 ± 30°, β = 178 ± 30°, γ = 54 ± 30°, ε = −155 ± 30°, ζ = −71 ± 30° as described by Chang and Tinoco (47) and Kieken et al. (17). Torsion angles χ (chi) were derived from the observation of intra-residue H6/8-H1′cross-peak volumes. All bases, except G12 and G11*, were restrained to anti-conformation (χ = −158 ± 30°). Torsion angles values ν0 to ν4 were derived from the analysis of COSY-DQF and TOCSY experiments. Nucleotides with no COSY and no TOCSY cross-peaks between H1′ and H2′ protons, i.e. all nucleotides except G11*, were restrained to the C3′-endo-conformation (ν0 = 6 ± 15°, ν1 = −25 ± 15°, ν2 = 37 ± 15°, ν3 = −37 ± 15°, ν4 = 21 ± 15°). This first set of structural restraints was used for structure calculations. Analysis of converged structures revealed that for U7 to G12 and C7* to A10*, classical A-helix ε angles values were observed. For G8 to G10 and C7* to A10*, classical A-helix ζ angles values were also observed. Thus, for U7 to G12 and C7* to A10* nucleotides, classical A-helix ε angles values (ε = −155 ± 30°) were added and for G8 to G10 and C7* to A10*, classical A-helix ζ angles values (ζ = 71 ± 30°) were also included. This second set of structural restraints was used for structure calculations. These constraints led to higher convergence efficiency with similar converged structures. In addition, in order to validate our structures, two additional calculations were run on the one hand including only ε angles values for U7 to G12 and C7* to A10* nucleotides, and on the other hand including only ζ angles values for G8 to G10 and C7* to A10*. Both structure calculations led to similar results as those obtained without these angle restraints.
Structure determination
Structures were calculated using CNS (cristallography and NMR system) torsion angle molecular dynamics (TAMD) protocol for nucleic acids using NOE and dihedral angle restraints (48,49). LNA nucleotides (C9* and A10*) were defined by modifying the appropriate nucleotides C and A, and atomic charges were determined according to those previously described (50). One hundred structures were generated from two randomized extended strands. The first stage consisted of a high-temperature torsion angle dynamics for 30 ps at 20 000 K with a van der Waals scale factor of 0.1. During the second stage, the molecules were cooled for 40 ps of torsion angle dynamics with a van der Waals scale factor increased from 0.1 to 1. In the third stage, molecules were submitted to a second slow cooling for 35 ps in Cartesian space where the van der Waals scale factor increased from 1 to 4. Finally, 10 Powell cycles of energy minimization of 300 steps each were done. The structures with zero violation on NOE distance (0.2 Å), dihedral (5°) and with the lowest energy were selected. Structures were visualized and were analysed with MOLMOL and PyMOL software packages (51,52).
Coordinates
Coordinates of 10 converged NMR structures have been deposited in the Protein Data Bank (PDB ID code is 2PN9). Coordinates of converged NMR structures calculated with no ε and ζ angles restraints for the loop–loop helix have also been deposited in the Protein Data Bank (PDB ID code is 2OOM).
UV melting experiments
Thermal denaturation of complexes was monitored on a Uvikon XL UV/visible spectrophotometer (BIOTEK) equipped with a 10-positions sample holder and a Peltier temperature control accessory. The experiments were performed at 1 μM final concentration of each RNA hairpin in 20 mM cacodylate buffer, containing 0.3 mM (or 3 mM) Mg2+, pH 7.3 at 20°C, in 200 μl micro-quartz cuvettes. Samples were overlaid with 300 μl of mineral oil to prevent evaporation at high temperature. An initial 30-min equilibrium time at 5°C was included prior to the temperature ramping. Denaturation of the samples was achieved by increasing the temperature at 0.4°C/min from 5 to 95°C and the UV absorbance was followed at 260 nm. The melting temperature (Tm) was determined as the maximum of the first derivative of the UV melting curves.
Surface plasmon resonance
Surface plasmon resonance experiments were performed on a BIAcore™ 3000 apparatus (Biacore AB, Sweden) as described previously (20). Briefly, binding kinetics were performed at 23°C in 10 mM phosphate buffer, pH 7.2, containing 20 mM sodium chloride, 140 mM potassium chloride, 0.3 mM magnesium chloride and 0.005% surfactant P20 (Biacore). Samples were injected at 20 μl/min at two different concentrations. Three independent experiments were performed for each run. The kinetic parameters, kon and koff, were determined assuming a pseudo-first-order model by direct curve fitting of the sensorgrams using the Bia-evaluation 4.1 software (Biacore) (Supplementary Data, Figure 1 and Table 1). The dissociation equilibrium constant, Kd, was calculated as koff/kon.
Table 1.
Melting temperature, Tm, of TAR–aptamer derivatives complexes
| Complexes | Tm(°C) | |
|---|---|---|
| 0.3 mM Mg2+ | 3 mM Mg2+ | |
| TAR/LR06(GA) | 44.5 ± 0.5 | 56.5 ± 0.5 |
| TAR/LM06(GA) | 42.2 ± 0.8 | 57.5 ± 3.5 |
| TAR/R06(GA) | 31.5 ± 0.0 | 47.3 ± 0.3 |
R06(GA) is the parent RNA aptamer. LR06(GA) and LM06(GA) are LNA/RNA and LNA/2′-O-methyl chimeric derivatives of R06(GA), respectively.
RESULTS AND DISCUSSION
In the following section, aptamers derived from the parent molecule R06(GA) are named according to their chemical modifications: L for LNA, R for RNA and M for 2′-O-methyl. The chimeric derivatives of R06(GA) with LNA/RNA or LNA/2′-O-methyl combinations are therefore termed LR06(GA) and LM06(GA), respectively.
Choice of the oligonucleotides for NMR studies
NMR structural studies were performed on a complex formed by a 16-nt TAR hairpin and the 16-nt chimera LR06(GA) with LNAs at positions C9* and A10* in the loop (Figure 1A). For TAR, the sequence corresponding to the top part of the viral RNA element contains an extra G•C pair at the bottom of the stem to obtain a better transcription yield. The aptamer derivative LR06(GA) was chosen according to previous work on LNA/2′-O-methyl chimeric derivatives that identified the chimera of strongest affinity for TAR. The presence of two LNA residues at positions C9* and A10* was shown to lead to a ligand of higher affinity for TAR than the originally selected RNA aptamer (33). Considering the difficulty to obtain milligram quantity of a 2′-O-methyl/LNA chimera [LM06(GA)], we choose to study by NMR the complex formed between TAR and the corresponding RNA/LNA chimera LR06(GA). Both are known to be RNA mimics. We verified that LR06(GA) displayed the same properties as LM06(GA) for binding to TAR. We first compared the stability of LR06(GA)/TAR and LM06(GA)/TAR complexes by thermal denaturation. Both complexes displayed similar melting temperatures (Table 1). The affinity of these two chimeras for TAR was then measured by surface plasmon resonance. The equilibrium constants are in the same order of magnitude: 0.27 ± 0.28 and 1.60 ± 0.34 nM (at 3 mM Mg2+) for LM06(GA) and LR06(GA), respectively. Finally, we checked by NMR that LR06(GA) or LM06(GA) chimeras generate kissing complexes of similar secondary structure (data not shown).
The RNA–RNA complex TAR/R06(GA) was also prepared to provide a reference NMR spectrum for analysing the influence of LNA residues on the base pairs involved in the loop–loop helix (Figure 1A and B).
Comparison of TAR/LR06(GA) and TAR/R06(GA) complexes
The loop–loop complexes were first characterized by thermal denaturation experiments monitored by UV absorption spectroscopy. Two melting transitions were observed for complexes between TAR hairpin and LR06(GA) or R06(GA) aptamers (data not shown). The higher transition (above 70°C) resulted from the melting of the hairpin stems while the lower transition resulted from the melting of the bimolecular complexes. The TAR/LR06(GA) complex (Tm = 44.5 ± 0.5°C) is more stable than the TAR/R06(GA) complex (Tm = 31.5 ± 0.5°C) (Table 1). Two LNA residues in the loop of the aptamer clearly increase the stability of the complex (ΔTm = 13°C). The affinity of R06(GA) and LR06(GA) for TAR was measured by surface plasmon resonance. LR06(GA) displays a higher affinity for TAR than R06(GA): Kd = 27.8 ± 3.4 and 355.0 ± 18.0 nM, respectively (Supplementary Table 1 and Figure 1).
We then compared by NMR the secondary structure of these two complexes (Figure 1C). Analysis of imino protons region points out subtle structural differences, in particular for the loop–loop helix, due to the presence of LNA residues in the aptamer loop. These results are described below.
Assignment of proton resonances
Assignment of TAR/R06(GA) kissing complex, based on analysis of NOESY spectra, had been previously reported (34). The secondary structure of the TAR/LR06(GA) complex (Figure 1A) was characterized by the analysis of 2D NOESY and 1H/15N HSQC experiments recorded in 90%/10% H2O/D2O. The imino proton spectrum of TAR exhibits five resonances showing the formation of a folded hairpin with 5-bp stem (data not shown). The imino proton spectrum of LR06(GA) indicates the formation of a folded hairpin too. Titration of TAR by the aptamer LR06(GA) was monitored in the imino region of 1D spectra. Each of the five imino proton resonances of TAR is shifted when the aptamer is added. The appearance of new resonances that were not observed for the free TAR or free aptamer with increasing aptamer concentration indicates the formation of new base pairs. All base-paired imino protons were assigned via sequential NOEs observed in 2D NOESY spectra. Analysis of H2O NOESY spectra supports the existence of three helical segments in TAR/LR06(GA) complex: two of them correspond to the stem of each hairpin and the third one is the helix that results from the loop–loop interaction. The complex between TAR and LR06(GA) is formed through 5 bp involving C6 to G10 of TAR, and C7* to G11* of LR06(GA) as shown in Figure 1A. The observation of the G5* imino proton (at 13.80 p.p.m. at 15°C) indicates that it is protected from exchange with H2O and that the G5*•A12* base pair is not in sheared conformation (Figure 1C). Assignment of base-paired imino protons of TAR was confirmed by the analysis of a 1H/15N HSQC experiment recorded on a complex formed by LR06(GA) and 13C/15N TAR (Figure 2A). Three adenine H2 protons were assigned to A3, A2* and A10* from the strong NOE observed between imino proton of A•U base pair and the H2 of adenine residue. The H2 proton of A12* was assigned from the NOE observed with the imino proton of G5* and was confirmed by inspection of experiments recorded in D2O.
Figure 2.
Assignment of proton, nitrogen and carbon resonances of TAR/LR06(GA)complex. (A) Imino proton region of the 1D spectrum (top) and the 1H/15N HSQC spectrum (bottom). (B) H6/8-H1′ region of a NOESY spectrum recorded at 15°C in 100% D2O with a mixing time of 200 ms. Dashed lines represent sequential assignment of H6/8-H1′. (C) 1H-13C HSQC spectrum showing the aromatic H8-C8, H6-C6 and H2-C2 correlations of TAR 13C-15N labelled complexed with LR06(GA).
Assignments of non-exchangeable protons were obtained using homonuclear and heteronuclear multidimensional experiments. As TAR was 13C/15N labelled, resonances from TAR and LR06(GA) were discriminated without ambiguity in 2D-NOESY spectra recorded in 100% D2O using 1H/13C HSQC experiment (Figure 2B and C). Sequential H6/8-H1′ connectivities observed in classical helix A were identified from G1 to C5, C13 to C16, C1* to G5*, A12* to G16*, C6 to A11 and U6* to G11*. Intermolecular connectivity between U*6 and C5 was also assigned. No H8-H1′ connectivity was observed for G12, except the connectivity A11(H8)-G12(H8). For LR06(GA), assignments were confirmed using 3D NOESY-NOESY experiment but only 67% of the protons were assigned because of the impossibility to 13C/15N label the LNA residues. Missing assignments correspond to sugar protons H3′, H4′ and H5′/H5″. For TAR, assignments were confirmed and completed using 3D HCCH-TOCSY and 3D NOESY-HMQC experiments.
NMR structure determination
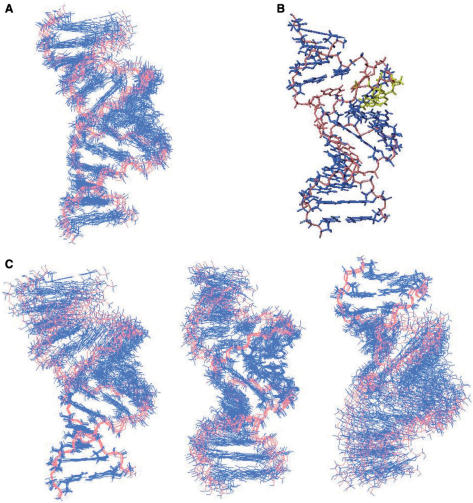
A total of 384 NOE distance restraints and 272 dihedral torsion restraints were derived from NMR data. Structures were calculated using CNS TAMD protocol for nucleic acids using NOE and dihedral angle restraints (48). The overall structure is well defined with a heavy atom root-mean-square deviation (r.m.s.d.) of 0.94 ± 0.09 Å for the 10 structures (Table 2, Figure 3A and B). The stems G1–G4/C13–C16, C1*–G4*/C13*–G16* present a typical A-type conformation. The central helix and the stem of LR06(GA) are less defined than the TAR stem, due to the non-availability of labelled LR06(GA) (Table 2, Figure 3C).
Table 2.
NMR restraints and statistics for the 10 converged structures
| Distance and dihedral restraints | |
|---|---|
| Intra-residue distance restraints | 141 |
| Inter-residue distance restraints | 159 |
| Hydrogen bonding distance restraints | 84 |
| Total distance restraints | 384 |
| Torsion angle restraints for sugar pucker | 185 |
| Backbone angle restraints | 87 |
| Total angle restraints | 272 |
| Total restraints | 656 |
| R.m.s.d. from experimental restraints | |
| Distance restraints (Å) | 0.0045 ± 0.0007 |
| Dihedral restraints (°) | 0.0499 ± 0.0160 |
| Deviation from idealized geometry | |
| Bond (Å) | 0.00300 ± 0.00006 |
| Angle (°) | 0.8241 ± 0.0099 |
| Impropers (°) | 0.3516 ± 0.0272 |
| R.m.s.d.a (Å) | |
| Heavy atom for all structures | 0.94 ± 0.09 |
| Backbone for all structures | 1.03 ± 0.10 |
| Heavy atom for stem helix (G1–C5, G12–C16) | 0.39 ± 0.12 |
| Backbone for stem helix (G1–C5, G12–C16) | 0.39 ± 0.14 |
| Heavy atom for central stem helix (C6–G10, C7*–G11*) | 0.60 ± 0.09 |
| Backbone for central stem helix (C6–G10, C7*–G11*) | 0.66 ± 0.09 |
| Heavy atom for stem helix (C1*–G4*, C13*–G16*) | 0.57 ± 0.17 |
| Backbone for stem helix (C1*–G4*, C13*–G16*) | 0.62 ± 0.18 |
aroot mean square deviation ± SD compared to the global average structure.
Figure 3.
Overall structure of TAR/LR06(GA) complex. (A) Superposition of 10 converged NMR structures. Bases are shown in dark blue and the backbone in salmon. (B) View of the average structure of TAR/LR06(GA) complex. LNAs residues are coloured in yellow, G5*•A12* and U6*•A11 in salmon. (C) The TAR stem region has been superimposed for the 10 converged structures (left). The central helix has been superimposed for the 10 converged structures (middle). The LR06(GA) stem region has been superimposed for the 10 converged structures (right).
Description of the loop–loop interaction
The loop–loop conformation is close to a A-type helix
The helix resulting from the loop–loop interaction is well defined with a heavy atom r.m.s.d. of 0.60 ± 0.09 Å for the 10 structures, compared to the average structure (Table 2, Figure 4A). The conformation of the loop–loop interaction involved in the complex formed by TAR and LR06(GA) is close to an A-type helix (Figure 4B and C). This conformation differs from that described by Chang and Tinoco for the complex formed by TAR and a rationally designed hairpin, termed TAR*(UA), with a complementary loop sequence 5′-UUCCCAGA-3′ (U and A residues closing the loop) (47). TAR*(UA) has a lower affinity for TAR than the R06(GA) aptamer (92.5 ± 5.2 nM versus 3.8 ± 5.2 nM) (18). In addition, the TAR/R06(GA) complex (Tm = 31.5 ± 0.5°C) is more stable than the TAR/TAR*(UA) complex (Tm = 18.4 ± 0.4°C) (53). The structure of the TAR/TAR*(UA) has been previously solved by NMR (PDB Id: 1KIS, 47). In contrast to TAR/LR06(GA), the loop–loop helix of TAR/TAR*(UA) displays a narrower minor groove compared to a canonical A-form helix (Figure 4D). This different conformation between the two complexes may result from two effects: the presence of the LNA residues in the loop on the one hand, and the presence of the G•A base pair instead of U•A loop closing pair on the other hand. These structural features might explain different behaviours of TAR*(UA) and LR06(GA).
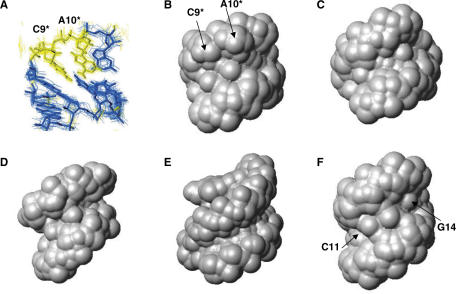
Figure 4.
Description of the loop–loop Interaction of TAR/LR06(GA) and comparison to LNA/RNA and RNA/RNA complexes. (A) View of the loop–loop interaction of TAR/LR06(GA) with LNA residues, C9* and A10*, coloured in yellow. View of the minor groove water accessible surface of (B) TAR/LR06(GA) loop–loop helix, (C) an A-type helix, (D) TAR/TAR*(UA) (1KIS), (E) LNA:RNA hybrid (1HOQ) and (F) SL1 HIV-1 dimer loop–loop helix (2F4X). The surfaces are generated using a water probe of 1.4 Å radius on MOLMOL software. PDB ID are indicated in brackets.
We then compared the loop–loop conformation of TAR/LR06(GA) with LNA-modified duplexes that have been investigated by NMR (32,50,54). Structural studies performed on LNA:DNA and LNA:RNA hybrids showed that increasing numbers of LNA in the hybrid resulted in an increased A-like character of the duplex: the minor groove width is progressively widened upon introduction of LNA modifications (Table 3) (32). The minor groove width can be characterized by the shortest distance between a given phosphate in one strand and phosphates in the opposite strand (55,56). It depends on the overall duplex conformation: B-type duplexes have a minor groove width of ∼5.7 Å whereas in A-type duplexes, the corresponding distance ranges from 10.0 to 12.0 Å (Table 3). NMR structural studies performed on LNA-modified DNA quadruplexes have pointed out that their minor grooves are larger than in their DNA counterpart (PDB Id: 2CHK, 2CHJ) (54). Similar results have been observed with LNA-modified DNA strand hybridized to RNA. As an example, in the structure of a RNA strand hybridized to a DNA containing three LNA residues (LNA3:RNA), the average minor groove width (9.6 Å) is larger than in the corresponding DNA:RNA hybrid (8.7 Å) [PDB Id: 1HHX, (32)]. In the fully modified LNA:RNA hybrid, the minor groove width is uniform along the helix from 9.7 to 10.3 Å (PDB Id: 1HOQ) (50) (Figure 4E). The loop–loop conformation of TAR/LR06(GA) is slightly different from this hybrid. The groove width is not uniform along the central helix with a phosphorus inter-strand distance varying from 10.13 to 12.15 Å (Table 3). A loop–loop interaction close to the one of TAR/LR06(GA) conformation has been recently described (17) (PDB Id: 2F4X). The loop–loop interaction of the RNA–RNA dimer formed by SL1 HIV-1Lai presents a geometry similar to that of TAR/LR06(GA) (Figure 4F). The groove width also ranges from 10.87 to 12.5 Å (Table 3). Thus, our structure shows that the introduction of two LNA nucleotides in a loop–loop context allows the hybridization with a RNA partner retaining normal Watson–Crick interactions, as shown by NMR data. The stacking pattern in the loop–loop helix is effectively observed: classical sequential connectivities and the inter-strand purine–purine stacking between A10* and G8 are present in NMR data.
Table 3.
Groove widths measured as the shortest inter-strand phosphorus distances minus 5.8 Å, which is the combined van der Waals radii of two phosphate groups (54)
| Number of LNA residues | Groove widths (Å) | |
|---|---|---|
| A-type helix (54) | 0 | 10–12 |
| B-type helix (54) | 0 | 5.7 |
| TGLGLT (2CHJ, 53) | 2 | 6.8 |
| TL4T (2CHK, 53) | 4 | 7.1 |
| DNA:RNA (32) | 0 | 8.7 |
| LNA1:RNA (1HHW, 32) | 1 | 8.9 |
| LNA3:RNA (1HHX, 32) | 3 | 9.6 |
| LNA:RNA (1HOQ, 50) | 9 | 9.7–10.3 |
| TAR/LR06(GA) (2PN9) | 2 | |
| Nucleotide step | ||
| C7*–C8* | 11.82 ± 0.40 | |
| C8*–C9* | 12.15 ± 0.36 | |
| C9*–A10* | 10.57 ± 0.47 | |
| A10*–G11* | 10.13 ± 0.52 | |
| SLI HIV-1 dimer (2F4X,17) | 0 | |
| Nucleotide step | ||
| C11–G12 | 10.87 ± 0.15 | |
| G12–C13 | 12.48 ± 0.06 | |
| C13–G14 | 12.53 ± 0.24 | |
| G14–C15 | 11.06 ± 0.16 |
PDB Id and reference are indicated in brackets. 5′-d(TGLGLT)-3′ and 5′-d(TL4T)-3′, with 2 and 4 LNA-modified G-residues respectively, form quadruplex structures. LNA1 is a LNA-modified DNA strand: 5′-d(CTGATATGC)-3′ where the LNA residue is underlined (32). LNA3 is also a LNA-modified DNA strand (5′-d(CTGATATGC)-3′), and LNA is the fully LNA-modified DNA strand (5′-CTGATATGC-3′) (32,50). LNA1, LNA3 and LNA are hybridized to the following RNA strand: 5′-GCAUAUCAG-3′.
G11*•C6 base pair is stabilized in TAR/LR06(GA) compared to TAR/R06(GA) complex
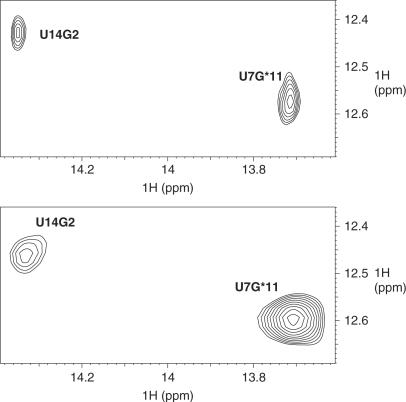
G11*•C6 is involved in the loop–loop interaction. As G11* is close to the two LNA residues C9* and A10*, we compared this base pair to its counterpart in the RNA complex TAR/R06(GA). In both complexes, G11*•C6 is a Watson-Crick base pair (Figure 1A and C). However, analysis of NMR data reveals differences. We have indeed noticed that G11* is more protected from solvent exchange in TAR/LR06(GA) than in TAR/R06(GA). In TAR/R06(GA), the signal of the imino proton of G11* overlaps the one of G14*. To compare the G11* in TAR/R06(GA) and in TAR/LR06(GA), we quantified the intensity of the correlation in NOESY spectra recorded in the same conditions for both complexes, using the imino–imino correlation between U14 and G2 as an internal reference for each complex. We clearly see that the imino proton of G11* in TAR/LR06(GA) exhibits stronger correlations than in TAR/R06(GA), as exemplified in Figure 5, for G11* and U7. Moreover, as the exchange rate of imino proton with the solvent is related to the dissociation constant of base pairs (57–59), this indicates that the G11*•C6 base pair is more stable in TAR/LR06(GA) than in TAR/R06(GA).
Figure 5.
Comparison of G11*•C6 base pair in TAR/R06(GA) and TAR/LR06(GA) complexes. Imino region of NOESY spectra recorded at 4°C in H2O/D2O 90%/10% of TAR/R06(GA) (top) and TAR/LR06(GA) (bottom).
Description of the structure at stem-loop junctions
G5*•A12* base pair is not in a sheared conformation
As demonstrated by NMR data, the G5*•A12* pair in TAR/LR06(GA) complex is not in sheared conformation (Figures 1C and 6A). The G5*•A12* is a non-canonical base pair in which N1 of A12* is hydrogen bonded to N1 of G5* (average distance of 2.94 ± 0.23 Å in the ensemble of 10 structures) and O6 of G5* is hydrogen bonded to N6 of A12* (average distance of 2.74 ± 0.11 Å in the ensemble of 10 structures). Moreover, NMR experiments recorded on the complex TAR/R06(GA) show that G5* is also protected from the exchange with the solvent (Figure 1C). The resonance corresponding to the imino proton of G5* is observed at 13.76 and 13.72 p.p.m. (at 4°C) for TAR/LR06(GA) and TAR/R06(GA), respectively. Thus, the G5*•A12* base pair adopts the same conformation in both complexes.
Figure 6.
Structure at the stem-loop junctions. (A) View of the G5*•A12* base pair in TAR/LR06(GA) complex. (B) View of A11 and U6* at stem-loop junction in TAR/LR06(GA) complex. (C) View of turns typically observed at the stem-loop junctions. TAR is represented in blue and the aptamer in salmon. Turns between C5 and C6 and between G5* and U6* in TAR/LR06(GA) complex (left). Turns between C5 and C6 and between U5* and U6* in TAR/TAR*(UA) complex (right) (47).
A11 and U6* pair deviates from a classical Watson–Crick base pair
The resonance corresponding to U6* imino proton cannot be safely assigned due to the absence of imino–imino correlation involving U6* in NOESY experiments with no magnesium. Moreover, the absence of correlation between the H2 proton of A11 and the imino proton of U6* indicates that this pair deviates from a classical Watson–Crick base pair. The analysis of NOESY spectra reveals that A11 and U6* are not involved in a reverse Hoogsteen base pair either. A11 exhibits sequential connectivities H8–H1′ and H8–H8 with G10 and G12, respectively, indicating that A11 is stacked. U6* exhibits an intermolecular and a sequential connectivity with C5 and U7*, respectively, indicating that U6* is stacked.
The melting temperature for TAR/LR06(GA) shows that the complex is stabilized when the concentration of magnesium is increased (Table 1). Thus, we examined the effect of the addition of magnesium on the structure. The chemical shifts and NOE patterns observed for the complex in the presence of 0.3 mM Mg2+ were not significantly affected (Supplementary Figure 2). At 3 mM Mg2+, results were similar with the exception of the appearance of the resonance corresponding to the imino proton of U6* (Supplementary Data, Figure 2). Two-dimensional NOESY experiment recorded in these conditions clearly points out that the imino proton of U6* exhibits a very weak correlation with the H2 proton of A11 in contrast to a classical Watson–Crick base pair (Supplementary Figure 3). This result and the chemical shift of the imino proton suggest that U6* and A11 might be engaged in a non-canonical U•A pair as described in other examples (60) (http://prion.bchs.uh.edu/bp_type). To test this hypothesis, a hydrogen bond between U6*(N3H) and A11(N1) was added for structure calculation. Analysis of the converged structure reveals that the A11 and U6* residues are involved in a U·A base pair that deviates from a classical Watson–Crick base pair (Figure 6B). N6 of A11 is hydrogen bonded to O4 of U6* (average distance of 3.01 ± 0.34 Å in the ensemble of 10 structures) and N3 of U6* is hydrogen bonded to N1 of A11 (average distance of 3.56 ± 0.13 Å in the ensemble of 10 structures). The average distance between U6*(H3) and A11(H2) in the ensemble of 10 structures is 4.05 ± 0.25 Å which explains the very weak correlation observed between those protons in NOESY spectra.
Stem-loop junctions present turns typically observed in ‘kissing’ complexes
Stems of TAR and LR06(GA) present a typical A-type conformation. The loop–loop helix creates turns at the stem-loop junctions between C5 and C6 and between G5* and U6* (Figure 6C). These structural features have been previously described in the structure of TAR/TAR*(UA) (47). We also observed short inter-phosphorus distance (4.82 ± 0.60 Å) between the 5′ phosphate group of C5 and the 5′ phosphate group of U7 of TAR in TAR/LR06(GA). In TAR/TAR*(UA), the corresponding distance was 5.25 ± 0.77 Å. In the turn involving G5*, inter-phosphorus distance between G5* and C7* is 6.93 ± 0.89 Å whereas this distance is 6.70 ± 0.90 Å in the structure of TAR/TAR*(UA) with U5* instead of G5* (47). One inter-backbone hydrogen bond, mediated via H2O molecules (61) between C5(H2′) and U6*(OP) might be formed. The average distance between C5(H2′) and U6*(OP) in the ensemble of 10 structures of TAR/LR06(GA) is 3.55 ± 0.50 Å.
CONCLUSION
We investigated the structure of a complex formed between the TAR RNA element of HIV-1 and a LNA/RNA chimera derived from an aptamer identified previously (7,8,33). Two important parameters account for the high stability of this complex.
First of all, LNAs at positions C9* and A10* in the aptamer loop induce an increased stability of the loop–loop helix. We have observed that the C6•G11* pair is more stable than the homologous pair in the RNA–RNA complex TAR/R06(GA). We have also noticed that the conformation of the loop–loop helix of TAR/LR06(GA) differs from the one described by Chang and Tinoco (47). The loop–loop geometry of TAR/LR06(GA) is similar to the one of a RNA–RNA dimer recently described (17). Indeed, the loop–loop dimer formed by SL1 HIV-1Lai is close to a canonical A-type helix, except at the stem-loop junctions (17). Thus, our structure shows that the introduction of two LNA nucleotides in a loop–loop context allows the hybridization with a RNA partner, retaining a normal Watson–Crick duplex framework and stabilizing adjacent pairs, with a loop–loop conformation close to an A-type helix. The increased stability due to the presence of LNA residues originates from the pre-organized geometry in a C3′-endo-conformation that likely provides an ideal geometry for the complex formation. LNAs would promote stacking interactions stabilizing the complex. This phenomenon has been previously described for 2′-deoxy-2′-fluoro-β-d-arabinonucleosides (2′F-ANA) (53). Thermodynamic parameters were measured on hybrids formed between 2′F-ANA and RNA or DNA. It was shown that the increased stability of the complex appeared to originate in conformational pre-organization of the fluorinated sugars and a favourable enthalpy of hybridization.
The second important structural feature is the formation of the non-sheared G5*•A12* base pair closing the loop of the aptamer. This provides additional stabilization at the helical junction compared to the UA combination present in the complex TAR/TAR*(UA) (47). The replacement of a pyrimidine cycle by a purine cycle increased the stacking effect. This accounts for the enhanced affinity of aptamers with G•A base pair for TAR over the rationally designed hairpin with U•A closing the loop (18) (Table 1). Our experimental results are in agreement with the molecular dynamics simulation performed by Beaurain et al. (62) on two RNA–RNA complexes derived from TAR/TAR*(UA). The authors used the TAR/TAR*(UA) NMR solution structure as the starting structure to model the complex with a GA loop closing combination. They replaced the U by a G and positioned the G and A residues in a cis Watson–Crick/Watson–Crick base pair according to preliminary NMR results.
Finally, in a previous work, the inhibitory effect of LNA derivatives was tested in a TAR-dependent double-luciferase HeLa cell reporter system (33). It was demonstrated that the aptamer LR06(GA) inhibited the TAR-dependent expression of a luciferase reporter gene whereas an aptamer derivative with four LNAs in the loop (LR062467) and that binds to TAR with a reduced affinity, had no effect (33). Previously, we also demonstrated that TAR/LR062467 and TAR/R06(GA) had different structures at the stem-loop junctions. Indeed, in the case of TAR/LR062467 U6* and A11 residues are involved in a classical Watson–Crick base pair and the non-sheared base pair G5*•A12* is not formed (34). Thus, the location of LNAs residues in the aptamer loop induces structural modifications at stem-loop junctions. These results unambiguously demonstrate that subtle structural changes in the loop–loop helix and at the stem-loop junctions of the kissing complex correlate with the observed biological effect.
The TAR RNA element, and more generally RNA structures constitute valid targets for designing agent of potential therapeutic interest (63). Aptamers that display a high specificity of recognition expand the virologist's tool base (64,65). Structural studies will bring additional information that will allow refining the ligands identified by SELEX.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
NMR experiments were recorded at the Metabolome Pole of Functional Genomics Platform at Bordeaux. We thank Mickaël Maucourt, Catherine Deborde and Dominique Rollin for providing access to the 500 MHz spectrometer located at INRA (UMR Biologie du Fruit), Villenave d’Ornon (FRANCE). We thank Sylvie Nonin-Lecomte for advices about CNS. Funding to pay the Open Access publication charges for this article was provided by INSERM (Institut National de la Santé et de la Recherche Médicale).
REFERENCES
- 1.Muesing MA, Smith DH, Capon DJ. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987;48:691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- 2.Isel C, Karn J. Direct evidence that HIV-1 Tat stimulates RNA polymerase II carboxyl-terminal domain hyperphosphorylation during transcriptional elongation. J. Mol. Biol. 1999;290:929–941. doi: 10.1006/jmbi.1999.2933. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout B. Multiple biological roles associated with the repeat (R) region of the HIV-1 RNA genome. Adv. Pharmacol. 2000;48:29–73. doi: 10.1016/s1054-3589(00)48003-8. [DOI] [PubMed] [Google Scholar]
- 4.Berkhout B, Silverman RH, Jeang KT. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 5.Karn J. Tackling Tat. J. Mol. Biol. 1999;293:235–254. doi: 10.1006/jmbi.1999.3060. [DOI] [PubMed] [Google Scholar]
- 6.Feng S, Holland EC. HIV-1 tat trans-action requries the loop sequence within TAR. Nature. 1988;334:165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- 7.Ducongé F, Toulmé JJ. In vitro selection identifies key determinants for loop-loop interactions: RNA aptamers selective for the TAR RNA element of HIV-1. RNA. 1999;5:1605–1614. doi: 10.1017/s1355838299991318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boiziau C, Dausse E, Yurchenko L, Toulmé JJ. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 1999;18:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- 9.Collin D, van Heijenoort C, Boiziau C, Toulmé JJ, Guittet E. NMR characterization of a kissing complex formed between the TAR RNA element of HIV-1 and a DNA aptamer. Nucleic Acids Res. 2000;28:3386–3391. doi: 10.1093/nar/28.17.3386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eguchi Y, Tomizawa JI. Complex formed by complementary RNA stem-loops and its stabilization by a protein: function of ColE1 Rom protein. Cell. 1990;60:199–209. doi: 10.1016/0092-8674(90)90736-x. [DOI] [PubMed] [Google Scholar]
- 11.Kim CH, Tinoco I. A retroviral RNA kissing complex containing only two G•C base pairs. Proc. Natl Acad. Sci. USA. 2000;97:9396–9401. doi: 10.1073/pnas.170283697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skripkin E, Paillart JC, Marquet R, Ereshmann B, Ereshmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muriaux D, Foose P, Paoletti J. A kissing complex together with a stable dimer is involved in the HIV-1Lai RNA dimerization process in vitro. Biochemistry. 1996;35:5075–5082. doi: 10.1021/bi952822s. [DOI] [PubMed] [Google Scholar]
- 14.Dardel F, Marquet R, Ehreshmann C, Ehresmann B, Blanquet S. Solution studies of the dimerization initiation site of HIV-1 genomic RNA. Nucleic Acids Res. 1998;26:3567–3571. doi: 10.1093/nar/26.15.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mujeeb A, Clever JL, Billeci TM, James TL, Parslow TG. Structure of the dimer initiation complex of HIV-1 genomic RNA. Nat. Struct. Biol. 1998;5:432–436. doi: 10.1038/nsb0698-432. [DOI] [PubMed] [Google Scholar]
- 16.Ennifar E, Walter P, Ehreshmann B, Ehresmann C, Dumas P. Crystal structures of coaxially stacked kissing complexes of the HIV-1 RNA dimerization initiation site. Nat. Struct. Biol. 2001;8:1064–1068. doi: 10.1038/nsb727. [DOI] [PubMed] [Google Scholar]
- 17.Kieken F, Paquet F, Brulé F, Paoletti J, Lancelot G. A new NMR solution structure of the SL1 HIV-1Lai loop-loop dimer. Nucleic Acids Res. 2006;34:343–352. doi: 10.1093/nar/gkj427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ducongé F, Di Primo C, Toulmé JJ. Is a closing “GA Pair” is rule for stable loop-loop RNA complexes? J. Biol. Chem. 2000;275:21287–21294. doi: 10.1074/jbc.M002694200. [DOI] [PubMed] [Google Scholar]
- 19.Darfeuille F, Arzumanov A, Gait MJ, Di Primo C, Toulmé JJ. 2′-O-methyl-RNA hairpins generate loop-loop complexes and selectively inhibit HIV-1 tat-mediated transcription. Biochemistry. 2002;41:12186–12192. doi: 10.1021/bi025974d. [DOI] [PubMed] [Google Scholar]
- 20.Darfeuille F, Arzumanov A, Gryaznov S, Gait MJ, Di Primo C, Toulmé JJ. loop-loop interaction of HIV-1 TAR RNA with N3′->P5′ deoxyphosphoramidate aptamers inhibits in vitro tat-mediated transcription. Proc. Natl Acad. Sci. USA. 2002;99:9706–9714. doi: 10.1073/pnas.122247199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Darfeuille F, Hansen JB, Orum H, Di Primo C, Toulmé JJ. LNA/DNA chimeric oligomers mimic RNA aptamers targeted to the TAR RNA element of HIV-1. Nucleic Acids Res. 2004;32:3101–3107. doi: 10.1093/nar/gkh636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jepsen JS, Sorensen MD, Wengel J. Locked nucleic acid: a potent nucleic acid analog in therapeutics and biotechnology. Oligonucleotides. 2004;14:130–146. doi: 10.1089/1545457041526317. [DOI] [PubMed] [Google Scholar]
- 23.Vester B, Wengel J. LNA (Locked Nucleic Acid): high-affinity targeting of complementary RNA and DNA. Biochemistry. 2004;43:13233–13241. doi: 10.1021/bi0485732. [DOI] [PubMed] [Google Scholar]
- 24.Karkare S, Bhatnagar D. Promising nucleic acid analogs and mimics: characteristic features and applications of PNA, LNA, and morpholino. Appl. Microbiol. Biotechnol. 2006;71:575–586. doi: 10.1007/s00253-006-0434-2. [DOI] [PubMed] [Google Scholar]
- 25.Kauppinen S, Vester B, Wengel J. Locked nucleic acid: high-affinity targeting of complementary RNA for RNomics. Handb. Exp. Phamacol. 2006:405–422. doi: 10.1007/3-540-27262-3_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh SK, Nielsen P, Koshkin AA, Wengle J. LNA (locked nucleic acids): synthesis and high-affinity nucleic acid recognition. J. Chem. Commun. 1998;4:455–456. [Google Scholar]
- 27.Bodensgaard K, Petersen M, Singh SK, Rajwanshi VK, Kumar R, Wengel J, Jacobsen JP. Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chemistry. 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 28.Petersen M, Nielsen CB, Nielsen KE, Jensen GA, Bondensgaard K, Singh SK, Rajwanshi VK, Koshkin AA, Dahl BM, et al. The conformations of locked nucleic acids (LNA) J. Mol. Recognit. 2000;13:44–53. doi: 10.1002/(SICI)1099-1352(200001/02)13:1<44::AID-JMR486>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Wengel J, Petersen M, Nielsen KE, Jensen GA, Hakansson AE, Kumar R, Sorensen MD, Rajwanshi VK, Bryld T, et al. LNA (locked nucleic acids) and the diastereoisomeric alpha-L-LNA: conformational tuning and high-affinity recognition of DNA/RNA targets. Nucleosides Nucleotides Nucleic Acids. 2001;20:389–396. doi: 10.1081/NCN-100002312. [DOI] [PubMed] [Google Scholar]
- 30.Vester B, Lundberg LB, Sorensen MD, Babu BR, Douthwaite S, Wengel J. Improved RNA cleavage by LNAzyme derivatives of DNAzymes. Biochem. Soc. Trans. 2004;32:37–40. doi: 10.1042/bst0320037. [DOI] [PubMed] [Google Scholar]
- 31.Elmen J, Thonberg H, Ljungberg K, Frieden M, Westergaard M, Xu Y, Wahren B, Liang Z, Orum H, et al. Locked nucleic acids (LNA) mediated improvements in siRNA stability and functionality. Nucleic Acids Res. 2005;33:439–447. doi: 10.1093/nar/gki193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen M, Bodensgaard K, Wengel J, Jacobsen JP. Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J. Am. Chem. Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- 33.Di Primo C, Rudloff I, Reigadas S, Arzumanov AA, Gait MJ, Toulmé JJ. Systematic screening of LNA/2′-O-methyl chimeric derivatives of a TAR RNA aptamer. FEBS Lett. 2007;581:771–774. doi: 10.1016/j.febslet.2007.01.047. [DOI] [PubMed] [Google Scholar]
- 34.Lebars I, Richard T, Di Primo C, Toulmé JJ. LNA derivatives of a kissing aptamer targeted to the trans-activating responsive RNA element of HIV-1. Blood, Cells, Mol. Dis. 2007;38:204–209. doi: 10.1016/j.bcmd.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 35.Kao C, Zheng M, Rudisser S. A simple and efficient method to reduce nontemplated nucleotide addition at the 3 terminus of RNAs transcribed by T7 RNA polymerase. RNA. 1999;5:1268–1272. doi: 10.1017/s1355838299991033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puglisi JD, Wyatt JR. Biochemical and NMR studies of RNA conformation with an emphasis on RNA pseudoknots. Methods Enzymol. 1995;261:323–350. doi: 10.1016/s0076-6879(95)61016-2. [DOI] [PubMed] [Google Scholar]
- 37.Goddard TD, Kneller DG. San Francisco: University of California; 2004. SPARKY 3. [Google Scholar]
- 38.Sklenar V, Piotto M, Leppik R, Saudek V. Gradient-tailored water suppression for [1]H-[15]N HSQC experiments optimized to retain full sensitivity. J. Magn. Reson. 1993;102:241–245. [Google Scholar]
- 39.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 40.Plateau P, Guéron M. Exchangeable proton NMR without base-line distorsion using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 41.Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chem. Phys. Lett. 1980;69:185. [Google Scholar]
- 42.Bax A, Davis DG. MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson. 1985;65:355–360. [Google Scholar]
- 43.Griesinger G, Otting G, Wuthrich K, Ernst RR. Clean TOCSY for proton spin system identification in macromolecules. J. Am. Chem. Soc. 1988;110:7870. [Google Scholar]
- 44.Derome A, Williamson M. Rapid-pulsing artifacts in double-quantum-filtered COSY. J. Magn. Reson. 1990;88:170–185. [Google Scholar]
- 45.Bax A, Clore GM, Gronenborn AM. 1H-1H correlation via isotropic mixing of 13C magnetization: a new three-dimensional approach for assigning 1H and 13C spectra of 13C-enriched proteins. J. Magn. Reson. 1990;88:425–431. [Google Scholar]
- 46.Clore GM, Bax A, Driscoll PC, Wingfield PT, Gronenborn AM. Assignment of the side-chain 1H and 13C resonances of interleukin-1 beta using double- and triple-resonance heteronuclear three-dimensional NMR spectroscopy. Biochemistry. 1990;29:8172–8184. doi: 10.1021/bi00487a027. [DOI] [PubMed] [Google Scholar]
- 47.Chang K-Y, Tinoco I., Jr The structure of an RNA “kissing” hairpin complex of the HIV TAR hairpin loop and its complement. J. Mol. Biol. 1997;269:52–66. doi: 10.1006/jmbi.1997.1021. [DOI] [PubMed] [Google Scholar]
- 48.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 49.Nonin-Lecomte S, Felden B, Dardel F. NMR structure of the Aquifex aeolicus tmRNA pseudoknot PK1: new insights into the recoding event of the ribosomal trans-translation. Nucleic Acids Res. 2006;34:1847–1853. doi: 10.1093/nar/gkl111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nielsen KE, Rasmussen J, Kumar R, Wengel J, Jacobsen JP, Petersen M. NMR studies of fully modified locked nucleic acid (LNA) hybrids: solution structure of an LNA:RNA hybrid and characterization of an LNA:DNA hybrid. Bioconjugate Chem. 2004;15:449–457. doi: 10.1021/bc034145h. [DOI] [PubMed] [Google Scholar]
- 51.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14:29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 52.DeLano WL. South San Francisco, CA, USA: DeLano Scientific LLC; 2005. The PyMOL Molecular Graphics System. http://www.pymol.org. [Google Scholar]
- 53.Wilds CJ, Damha MJ. 2′-Deoxy-2′-fluoro-b-D-arabinonucleosides and oligonucleotides (2′F-ANA): synthesis and physicochemical studies. Nucleic Acids Res. 2000;28:3625–3635. doi: 10.1093/nar/28.18.3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nielsen JT, Arar K, Petersen M. NMR solution structures of LNA modified quadruplexes. Nucleic Acids Res. 2006;34:2006–2014. doi: 10.1093/nar/gkl144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pleij CWA, Rietveld K, Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985;13:1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haasnoot CAG, De Bruin SH, Hilbers CW, Van der Marel GA, Van Boom H. Loopstructures in synthetic oligonucleotides. Hairpin stability and structure as a function of loop elongation. Proc. Int. Symp. Biomol. Struct. Interactions, Suppl. J. Biosci. 1985;8:767–780. [Google Scholar]
- 57.Leroy JL. L’échange des protons imino: une sonde du mouvement d’ouverture des bases et de la structure des acides nucléiques. Regard sur la Biochimie. 1990;5:57–65. [Google Scholar]
- 58.Nonin S, Jiang F, Patel DJ. Imino proton exchange and base-pair kinetics in the AMP-RNA aptamer complex. J. Mol. Biol. 1997;268:359–374. doi: 10.1006/jmbi.1997.0986. [DOI] [PubMed] [Google Scholar]
- 59.Nonin S, Leroy JL, Guéron M. Terminal base pairs of oligodeoxynucleotides: imino proton exchange and fraying. Biochemistry. 1995;34:10562–10659. doi: 10.1021/bi00033a041. [DOI] [PubMed] [Google Scholar]
- 60.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA-solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 61.Hennig M, Williamson JR. Detection of N-H … N hydrogen bonding in RNA via scalar couplings in the absence of observable imino proton resonances. Nucleic Acids Res. 2000;28:1585–1593. doi: 10.1093/nar/28.7.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Beaurain F, Di Primo C, Toulmé JJ, Laguerre M. Molecular dynamics reveals the stabilizing role of loop closing residues in kissing interaction: comparison between TAR-TAR* and TAR-aptamer. Nucleic Acids Res. 2003;31:4275–4284. doi: 10.1093/nar/gkg467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toulmé JJ, Di Primo C, Moreau S. Modulation of RNA function by oligonucleotides recognizing RNA structure. Prog. Nucleic Acid Res. Mol. Biol. 2001;69:1–46. doi: 10.1016/s0079-6603(01)69043-3. [DOI] [PubMed] [Google Scholar]
- 64.James W. Aptamers in the virologists'toolkit. J. Gen. Virol. 2007;88(Pt 2):351–364. doi: 10.1099/vir.0.82442-0. [DOI] [PubMed] [Google Scholar]
- 65.Held DM, Kissel JD, Patterson JT, Nickens DG, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front. Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.