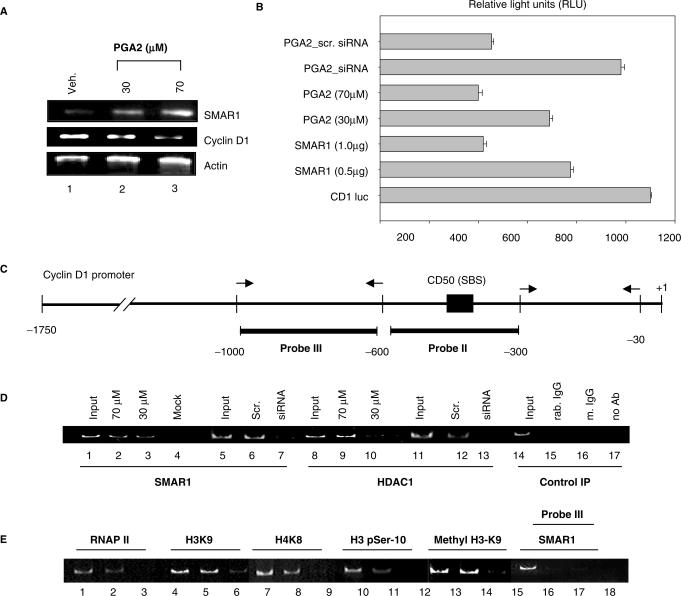
Figure 5.
Responsiveness of SMAR1 5′ UTR is essential for exerting its transcriptional modulatory function. (A) RT–PCR analysis of SMAR1, Cyclin D1, and β-actin transcript levels in PGA2-treated (lanes 2 and 3 at 30 and 70 μM PGA2) or vehicle-treated MCF-7 cells (lane 1). Actin was used as loading control (lower panel). (B) Luciferase activity from Cyclin D1 promoter was studied after transfection with the indicated plasmids, siRNA or PGA2 treatment and relative light units were plotted in the bar graph. Experiments were performed as three independent sets and error bars denote the mean deviation. From below, Bar 1 denotes the luciferase activity of CD1 promoter. Bars 2 and 3 denote the repression of Cyclin D1 by the intact UTR containing SMAR1 while bars 4 and 5 indicate down regulation of Cyclin D1 by the 30 and 70 μM PGA2. Use of SMAR1 specific siRNA and PGA2 treatment shows inefficient downregulation of Cyclin D1 luciferase activity. (C) Schematic representation of Cyclin D1 promoter and SMAR1-binding site (SBS) in probe II region. (D) Chromatin immunoprecipitation was performed as described in ‘Material and Methods’ section. Recruitment of SMAR1 (lanes 2 and 3) and HDAC1 (lanes 9 and 10) on probe II region of Cyclin D1 promoter was studied upon PGA2 treatment in MCF-7 cells using antirabbit SMAR1 and antimouse HDAC1 antibody. Endogenous SMAR1 was depleted using SMAR1 siRNA and studied for recruitment of SMAR1 (lane 7) and HDAC1 (lane13) in MCF-7 cells treated with PGA2 (70 μM). Scrambled siRNA (scr) was used as negative control that did not alter SMAR1 or HDAC1 recruitment (lanes 6 and 12, respectively). Chromatin immunoprecipitation using mouse IgG, rabbit IgG or no antibody are used as negative control (lanes 15–17). Reverse cross-linked sheared genomic DNA is used as positive control (input) for amplification (lanes 1, 5, 8, 11 and 14). Transfection of empty vector was performed as mock control (lane 4). (E) To study the status of histone modifications at Cyclin D1 promoter upon PGA2 treatment, ChIP assays were performed using antibodies against RNA pol II, acetyl H3K9, acetyl H4K8, H3 p-ser10 and monomethyl H3K9 and in MCF-7 cells untreated (lanes 2, 5, 8, 11 and 14) or treated with 70 μM PGA2 (lanes 3, 6, 9, 12 and 15) and Probe II region of Cyclin D1 promoter was amplified from the immunoprecipitated DNA. Reverse cross-linked sheared genomic DNA is used as positive control (input) for amplification (lanes 1, 4, 7 and 10). Probe III region was used as control to verify the specificity of SMAR1 binding to the promoter region.