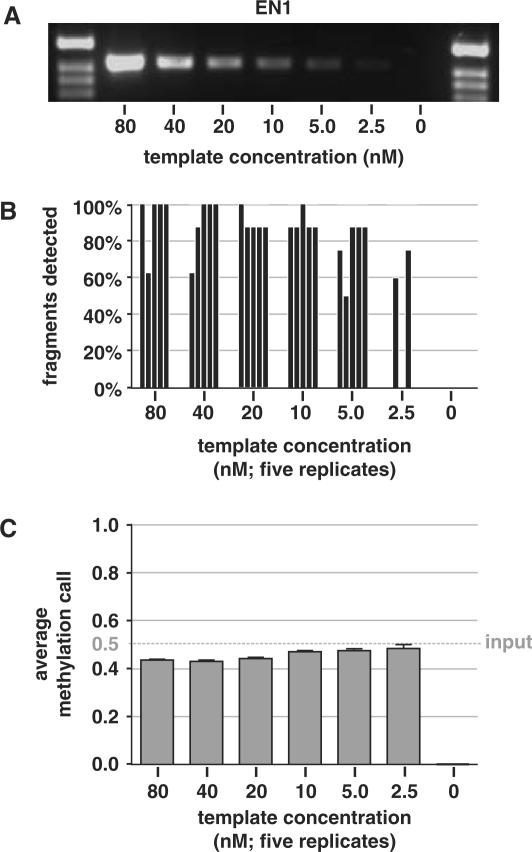
Figure 3.
Template dilution test for transcription and subsequent MALDI-TOF analysis. The minimal amount of PCR template required for accurate quantitation of DNA methylation was determined. (A) A twofold dilution series was prepared from T7-tagged PCR amplifications of EN1 CpG island (50/50% mixture of fully unmethylated and fully methylated PCR product), ranging from 80 to 2.5 nM, and 5 μl of each dilution was visualized on an agarose gel. Adjacent marker lanes contain 500 ng pBR322 digested with HinFI. (B) Two microliters of the dilution series was used in the transcription-RNase A digestion reaction and subjected to MALDI-TOF MS analysis, in five technical replicates. The ratio of detected versus expected fragments with CpG sites was determined in all spectra and are presented as individual bars for each analysis. (C) The average methylation calls ratios calculated from the detected peaks in the spectra are shown as means + SEM. The dotted horizontal line represents the actual methylation level used as input. Similar template dilution studies for SCTR and INHBB CpG island regions are given as Supplementary Data files.