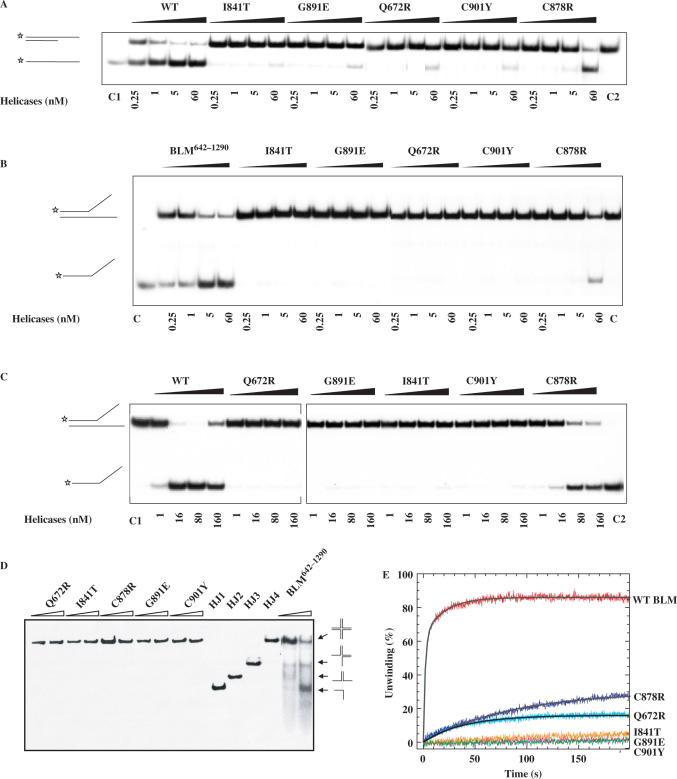
Figure 5.
DNA unwinding activity of wild-type and mutant BLM proteins. (A−C) Helicase activities revealed by radiometric assay using duplex DNA and forked DNA. 5′-32P-labelled DNA substrate (1 ng) was incubated at 37°C with wild-type and the various mutants in helicase assay buffer. The reactions were terminated after 30 min and the samples were analysed by electrophoresis in a 12% non-denaturing polyacrylamide gel. The concentrations of the proteins were between 0.25 and 60 nM as indicated at the bottom of the gel panel, where C1 and C2 represent labelled ssDNA and partial duplex DNA substrate, respectively. (D) Unwinding of the Holliday junction by BLM642–1290 and the all mutants. Reaction mixtures (20 µl) in unwinding buffer contain 1 nM DNA substrate, 1 mM ATP and the indicated amounts of the proteins. The four-way DNA junction was constructed from oligonucleotides HJ1, HJ2, HJ3 and HJ4. Reactions were carried out and analysed as described for A. HJ1, HJ2, HJ3 and HJ4 represent single-stranded, two-stranded, three-stranded and four-way junction substrate, respectively. (E) DNA unwinding activity by stopped-flow DNA unwinding assay. As described in the ‘Material and Methods’ section, 1 nM 56:16-mer DNA substrate was pre-incubated with 20 nM helicase for 5 min at 25°C. The unwinding reaction was initiated by mixing with 1 mM ATP. The fluorescence emission of fluorescein at 520 nm (excited at 492 nm) was monitored. The fraction of DNA unwound was obtained by normalization of the fluorescence signal. The solid curves are the best fit of the data to Equation (1) and the values of A1 (A2) and kobs,1 (kobs,2) are given in Table 2.