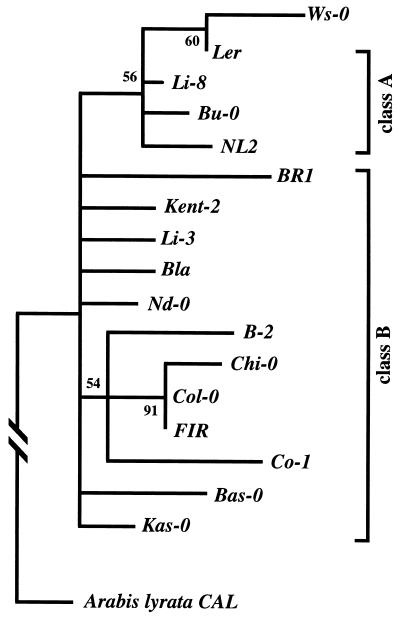
Figure 2.
Gene genealogy of the Arabidopsis CAL alleles based on maximum parsimony analysis. The distinctions between class A and B alleles are based on the pattern of shared polymorphisms. The numbers next to the nodes are the bootstrap values after 500 bootstrap replicates of the data; all nodes with <50% support are collapsed in the tree. Ten equally parsimonious trees (length = 528 steps) were obtained from a heuristic analysis; these trees were all congruent with the bootstrap results. The consistency index is 0.926, with the A. lyrata CAL orthologue as the outgroup. The genealogy is based on the sequence from exon 2 to the 3′ untranslated region. If only the recombinant exon 7 and flanking intron sequences are used, the Ws and Kent-2 alleles group together in the phylogeny (see text).