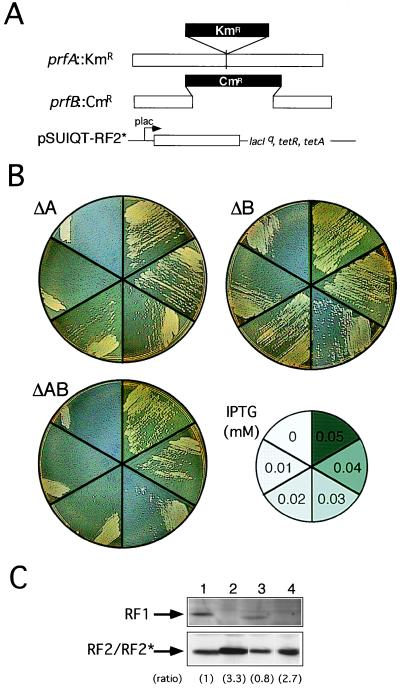
Figure 2.
Replacement of RF1 and RF2 function with RF2* by chromosomal gene disruption. (A) Disruption of RF1 (prfA) and RF2 (prfB) genes by KmR and CmR cassettes, respectively, on the E. coli chromosome. RF2* was cloned in plasmid pSUIQT so as to be expressed by the addition of IPTG. (B) Viability of pSUIQT-RF2* transformants of RF1 knockout (ΔA, prfA∷KmR), RF2 knockout (ΔB, prfB∷CmR), and RF1/RF2 double knockout (ΔAB, prfA∷KmR prfB∷CmR) strains. Cells were grown at 37°C on LB agar plates containing different IPTG concentrations as indicated. Cell viability can be judged by single colony formability, not by appearance of a cell zone (caused by cell density effect carrying over some IPTG) at the top of the cell suspension streak. (C) Western immunoblot analysis of the level of RF2* required for suppression of prfA and/or prfB knockout strains. Equal amounts (10 μg of bulk protein) of cell lysates were analyzed by SDS/PAGE and subjected to immunoblot staining with anti-RF1 (Top) and anti-RF2 (Bottom) antibodies. Lanes: 1, E. coli W3110 (nontransformant control); 2, pSUIQT-RF2* transformant of prfA∷KmR disruptant (0.04 mM IPTG); 3, pSUIQT-RF2* transformant of prfB∷CmR disruptant (0.01 mM IPTG); and 4, pSUIQT-RF2* transformant of prfA∷KmR prfB∷CmR double disruptant (0.04 mM IPTG). Lanes 2–4 represent RF2* synthesized with the minimal concentrations of IPTG sufficient to restore viability of prfA and/or prfB knockout strains. Relative intensities of immunoblots compared with that of lane 1 are shown in parentheses.