Figure 11. Multiple sequence alignment and secondary structure predictions of Soc.
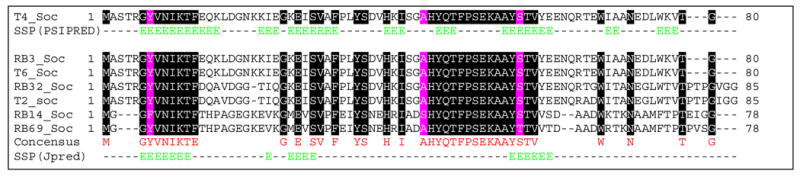
Seven T4 Soc-like sequences were aligned by ClustalW (www.ebi.ac.uk/clustalw/) and JPRED (www.compbio.dundee.ac.uk/~www-jpred/) to generate multiple sequence alignment and secondary structure predictions, respectively (accession numbers: T4_Soc: P03715; RB32_Soc: YP_802970; T2_Soc: AAK66983; RB69_Soc: NP_861717. Soc sequences of RB3, T6, and RB14 were from Tulane T4-like genome website, http://phage.bioc.tulane.edu/). The numbers on the left and right of sequence correspond to the number of aa in the respective Soc sequence. Identical aa are highlighted in black, and conserved aa, in pink. For secondary structure predictions, each Soc sequence was submitted in FASTA format to ClustalW to generate a multiple sequence alignment. The output file was submitted to the Jpred server to produce a secondary structure prediction (ssp) based on the consensus sequence. The secondary structure prediction of T4 Soc sequence was generated using PSIPRED (http://bioinf.cs.ucl.ac.uk/psipred/); E = β strand (green); dashes indicate no predicted structure.
