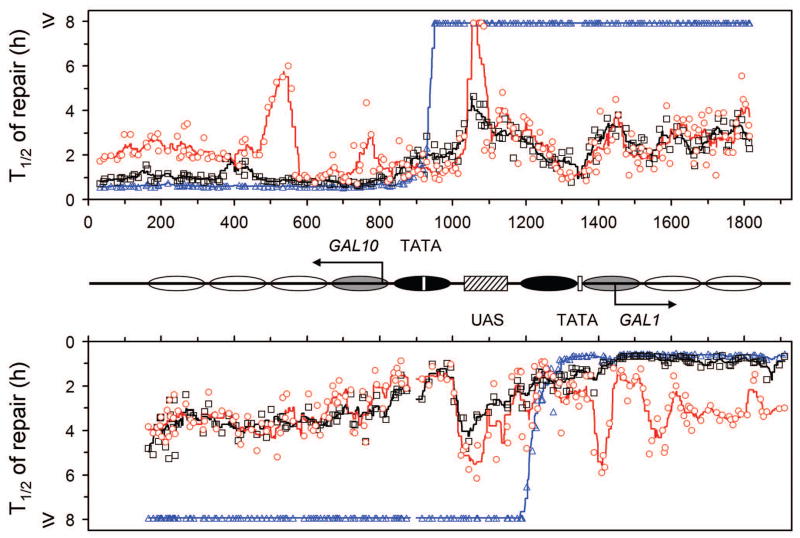
Fig. 5. Plot showing NER in the galactose induced GAL1-10 genes.
The upper and lower panels represent the top (TS for GAL10 and NTS for GAL1 genes) and bottom strands, respectively. Between the panels is a schematic diagram of the GAL1-10 region, where ovals denote the locations of nucleosomes, with the extent of shading reflecting the variability of positioning (darkest, least variable; lightest, most variable) [34]. Abscissa values are nucleotide positions numbered from a site that is 810 nucleotides downstream of the major transcription start site of the GAL10 gene. Arrows indicate the major transcription start sites. Open vertical bars denote TATA boxes of the GAL10 and GAL1 genes, and hatched box denotes the common UAS of the two genes. Individual symbols in the plot represent time (in hours) required for repairing 50% (T1/2) CPDs at individual sites of the GAL1-10 genes. Black squares, blue triangles and red circles represent wild type, rad16 and rad26 cells, respectively. Black, blue and red lines in the plot are smoothed T1/2 values for wild type, rad16 and rad26 cells, respectively. Smoothing was carried out by averaging the individual T1/2 values at continuous intervals of 40 nucleotides, where the 40 nucleotide brackets were ramped along the DNA by one nucleotide.