Abstract
A novel protein family, designated hereafter as RNase κ (kappa) family, has been recently introduced with the characterization of the specific Cc RNase, isolated from the insect Ceratitis capitata. The human ortholog of this family consists of 98 amino acids and shares > 98% identity with its mammalian counterparts. This RNase is encoded by a single-copy gene found to be expressed in a wide spectrum of normal and cancer tissues. The cDNA of the human ribonuclease has been isolated and subcloned into a variety of prokaryotic expression vectors, but most efforts to express it caused a severe toxic effect. On the other hand, the expression of the human RNase by the use of the methylotrophic yeast Pichia pastoris system resulted in the production of a highly active recombinant enzyme. Using a 30-mer 5′-end-labeled RNA probe as substrate, the purified enzyme seems to preferentially cleave ApU and ApG phosphodiester bonds, while it hydrolyzes UpU bonds at a lower rate. Based on amino acid sequence alignment and substrate specificity data, as well as the complete resistance of the recombinant protein to the placental ribonuclease inhibitor, we concluded that the human RNase κ is a novel endoribonuclease distinct from other known ribonucleases.
Accession No. AM746459
INTRODUCTION
RNA molecules are endowed with a very special role in the mechanism of gene expression and therefore in cell differentiation. A very wide spectrum of enzymes known as RNases catalyzes the degradation of polymeric RNA and participates in a wide range of biological events in both the intracellular and extracellular level (1,2). In recent years a focused study on RNases has led to the identification of novel ribonucleolytic enzymes in both prokaryotic and eukaryotic cells, even though their biological role in most cases remains undefined (2).
The RNase A superfamily has been a subject of intense biochemical study for many decades. Dozens of novel members of the RNase A superfamily have been identified in multiple vertebrate species and their biochemical properties as well as their biological roles have been intensively studied (3–14). Among the interesting features of this superfamily is its dynamic evolutionary history, which involved a series of gene duplications and advantageous amino acid substitutions.
Human RNases have been the epicenter of the interest in RNases for a long time. So far eight members of the RNase A superfamily have been studied in humans and despite the fact that they have been fully characterized in the biochemical level their in vivo activity remains obscure (13). On the other hand human exoribonucleases have been studied less extensively, but have been shown to exhibit key roles in the processing and maturation of human RNA molecules. The members of the RNase A superfamily share a relatively high level of homology and their genes are located on human chromosome 14. In addition to their catalytic activity, members of this superfamily have been shown to possess important biological actions other than a simple digestive role. As examples, human RNase 5 (angiogenin) promotes blood vessel formation, while human RNase 2 (eosinophil-derived neurotoxin, EDN) and human RNase 3 (eosinophil cationic protein, ECP) exert neurotoxic, immunosuppressive and antiviral activities (15–17). In a recent publication, five new members of the RNase A superfamily have been identified by in silico analysis in humans, even though some of them seem to lack the catalytic amino acids and therefore do not bear ribonucleolytic activity (18).
In this work, we present the molecular cloning and characterization of a novel human endoribonuclease, designated as human RNase κ, which belongs to a recently identified protein family. The first member of this family that has been studied is Cc RNase, isolated from the insect Ceratitis capitata (19). Cc RNase is a small, thermolabile ribonuclease consisting of 95 amino acids, which degrades poly (U), poly (C) and rRNA. The human ortholog was expressed in bacteria and yeast and its study revealed that the recombinant protein displays ribonucleolytic activity, as also was observed in the case of Cc RNase.
MATERIALS AND METHODS
Materials
Oligonucleotide synthesis and sequence analysis were performed by MWG Biotech (Ebersberg, Germany). Restriction enzymes, T4 DNA ligase and T4 polynucleotide kinase were purchased from New England Biolabs. Plasmid preparation kits and Protino His-Bind resin were from Macherey-Nagel. All bacterial expression vectors and bacterial strains, the anti-His tag monoclonal antibodies and the horseradish peroxidase conjugated goat anti-mouse IgG were purchased from Novagen. rRNA was extracted from rat liver polysomes as described previously by Rampias et al. (19). KM71 Pichia pastoris host cells and the yeast expression vector pPICZαA were purchased from Invitrogen. Multiple Tissue Northern Blot membrane was purchased from Clontech (#7760-1). Torula yeast RNA, the synthetic substrates poly (C) and poly (U) and the alkaline phosphatase conjugated goat anti-chicken IgG were obtained from Sigma (St Louis, MO/USA). Marker proteins for molecular weight estimations in SDS–PAGE were obtained from Fermentas. All other reagents used were of analytical grade and obtained from Merck.
Production of a specific polyclonal antibody against human RNase κ
The synthetic peptide AVLIEDVPFTEKDFE, corresponding to human RNase κ deduced amino acid sequence residues 38–52, was used for the preparation of the custom anti-human RNase κ specific polyclonal antibody. Peptide synthesis and antibody production was performed by Sigma-Genosys. Briefly, the synthesized peptide was purified by HPLC, conjugated to KLH and the conjugated synthetic peptide was used for the immunization of Rhode Island Red Chickens over a 10-week immunization protocol.
Isolation of the cDNA clone encoding human RNase κ
One microgram of poly(A)+ RNA from human placenta was reverse transcribed using the SMART PCR cDNA synthesis kit (Clontech Laboratories) as specified in the manufacturer's manual. In order to clone the human RNase κ cDNA, a PCR reaction was performed using the FH (5′- GCT TGC ACC TCG GCG AT-3′) and RH (5′-GAA GGG ATT CAG TCT CTC GC-3′) primers, which anneal at the 5′ and 3′ sequences of the untranslated regions of the human RNase κ mRNA, respectively. The PCR product was cloned into the pCR™ 2.1 cloning vector (Invitrogen) and sequenced in both directions.
Construction of the expression plasmids
Amplification of the open reading frame (ORF) of the human RNase κ was carried out using the proofreading Fast Start Taq DNA polymerase (Roche) and primers specific to the 5′ (starting at the ATG initiator codon) and 3′ ends of the human RNase κ cDNA based on nucleotide sequence (Accession No AM746459). The primers used were designed so that the amplified DNA would contain EcoRI and XbaI restriction endonuclease sites at its 5′ and 3′ ends, respectively, in order to facilitate subsequent subcloning. The primer sequences were as follows: sense, 5′- GAA TTC ATG GCG TCG CTC CTG TGC-3′; antisense, 5′- TCT AGA GCG CGC ACC ATG TAT TCC TTG-3′. The amplified cDNA fragment was first cloned into pCR™ 2.1 vector and the EcoRI–XbaI DNA fragment was purified and subcloned into the pPICZα A P. pastoris expression vector, creating the pαA construct. Using slightly modified sense and antisense primers containing different restriction sites, the human RNase κ cDNA was cloned into the prokaryotic expression vectors pET 15b (XhoI–BamHI insert), pET 20b (NcoI–XhoI insert), pET 29b (NspV–XhoI insert), pET 39b (NspV–XhoI insert), pET coco-1 (NheI–HindIII insert) and pSCREEN-1b(+) (NspV–XhoI insert). All constructs were confirmed by DNA sequencing.
Transformation of P. pastoris and selection of RNase-producing clones
Transformation of P. pastoris KM71 host cells was performed by electroporation as described by the manufacturer's manual. Briefly, SacI linearized pαA construct was electroporated with KM71 cells in a 0.2 ml sterile electroporation cuvette at 1.5 kV (Gene pulser, Bio-Rad). Cells were then plated onto yeast extract peptone dextrose sorbitol (YPDS; 1% yeast extract, 2% peptone, 2% glucose, 1 M sorbitol) agar plates containing 100, 200 and 500 g/ml Zeocin (Invitrogen), respectively and incubated at 30°C for 2–3 days until single colonies were formed. Screening for gene replacement of the construct by homologous recombination at the AOX1 locus, was confirmed by PCR analysis of chromosomal DNA using the 5′ AOX1 and 3′ AOX1 universal primers as described in the manufacturer's manual.
For the detection of the RNase-producing strains, a number of recombinant colonies was inoculated in 10 ml-buffered complex glycerol medium (BMGY; 1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4 × 10−5% biotin, 1% glycerol) and incubated at 30°C in a shaking incubator (250 r.p.m.). The cultures were centrifuged and the cells were re-suspended in appropriate volume of buffered complex methanol medium (BMMY; 1% yeast extract, 2% peptone, 100 mM potassium phosphate, pH 6.0, 1.34% YNB, 4 × 10−5% biotin, 0.5% methanol) so that OD600 was 2. The cultures were maintained for 4 days and supplemented daily with 0.5% v/v methanol. One milliliter of each culture was harvested every day; the supernatants were clarified by centrifugation (10 min, 10 000g, 4°C) and stored at −80°C until tested. The presence of recombinant RNase in the supernatants was detected by Dot Blot analysis using the anti-His tag monoclonal antibody as well as by their ability to cleave rRNA.
Large-scale production of recombinant human RNase κ was performed under similar conditions using the colony that rendered the best yield in the small-scale experiments.
Production and purification of recombinant human RNase κ in yeast
One hundred milliliters of extracellular medium obtained 2 days after the induction were concentrated about 20 times by ultrafiltration in a diaflow pressure cell with an YM-10 membrane. The concentrated material was diluted 1:4 with IBW buffer consisting of 50 mM sodium phosphate–NaOH pH 8.0, 300 mM NaCl and loaded onto a 1 ml Protino His-bind Resin column (Macherey-Nagel). After washing with the equilibration IBW buffer, the recombinant protein was eluted with IBW buffer containing 250 mM imidazole. The fractions containing the purified human RNase κ were pooled and the purified protein was dialyzed overnight against 20 mM Tris–HCl pH 8.0.
Protein purity was assessed by SDS–PAGE according to Laemmli (20). Polyacrylamide gels were either stained with Coomassie blue or electroblotted onto a nitrocellulose membrane as described by Towbin et al. (21). The recombinant protein was immunodetected using either an anti-His tag monoclonal antibody (1000-fold dilution) or a polyclonal antibody raised in chicken against a specific oligopeptide of the human RNase κ. Protein concentration was determined using a Bio-Rad (Richmond, CA, USA) protein assay kit based on the Bradford dye-binding assay (22). Calibration curves were constructed using bovine serum albumin as standard. Protein identification employing analysis of peptides resulting from the enzymatic digestion of the human RNase κ by LC-ESI-MS and MALDI-MS was performed by the Mass Spectrometry Lab of the Institute of Biochemistry and Biophysics of the Polish Academy of Sciences.
Expression of the human RNase κ in bacteria
Overnight cultures of several types of Escherichia coli host cells [Tunner(DE3), BL21(DE3), BL21(DE3)plysS, Rosetta(DE3), Rosetta(DE3)plysS] harboring the recombinant expression vectors mentioned above, were diluted 1:50 in LB or M9 medium supplemented with the appropriate antibiotics in each case. The effect of the recombinant protein expression on cell growth was investigated as described by Rosenberg (23). The purification of the recombinant human RNase κ, which had been expressed in BL21(DE3)plysS cells transformed with the recombinant pSCREEN-1b(+) vector, was achieved following the purification protocol employed in the case of Cc RNase (19).
Assay for RNase activity
RNase activity of the recombinant protein against torula yeast RNA (Sigma) or various single-stranded polyribonucleotides was determined by the production of perchloric acid-soluble material according to the method of Slifman et al. (24), with slight modifications. The reaction was performed at 37°C in a total volume of 50 μl, containing 50 μg of the various substrates, in 20 mM Tris–HCl pH 8.0 and the appropriate amount of recombinant protein. The reaction was stopped by addition of 0.2 ml ice-cold 7% perchloric acid containing 0.1% uranyl acetate and acid-soluble ribonucleotides remaining in the supernatant fraction after centrifugation were quantitated spectrophotometrically at 260 nm. The amount of enzyme used in the assays was selected such that the rate of hydrolysis would be linear in the time range examined.
Activity-staining gels
Recombinant human RNase κ activity towards poly (U) was also assayed by activity staining gel (25). The purified protein was diluted in sample buffer devoid of any reducing agent and analyzed in a 15% SDS polyacrylamide gel, containing 0.25 mg/ml poly (U) as substrate. Following electrophoresis, SDS was removed from the gel by washing (2 × 10 min) with 10 mM Tris–HCl buffer (pH 7.6) containing 2-propanol (20% v/v). Proteins were renatured by washing the gel twice with 10 mM Tris–HCl buffer (pH 7.6). The gel was then incubated in a buffer of 0.1 M Tris–HCl pH 7.6 for 3 h, to allow the degradation of the substrate to take place. The gel was stained with 10 mM Tris–HCl buffer (pH 7.6) containing toluidine blue (0.2% w/v) and distained with 10 mM Tris–HCl buffer (pH 7.6). Toluidine blue stains high-Mr nucleic acids leaving regions in the gel that contain ribonucleolytic activity appear as a clear band in a blue background.
Base specificity of the human RNase κ
The specific cleavage sites of the human RNase κ were determined by incubation of the ribonuclease with a 5′-[γ-32P] ATP-labeled synthetic 30-mer oligoribonucleotide of known sequence 5′-CCCCGAUUUUAGCUAUCUGGGUUCAACUUG-3′. Human RNase κ reactions were performed in mixtures containing 20 mM Tris–HCl pH 8.0. The 1 nt ladder was prepared by alkaline hydrolysis of the 30-mer 5′-end-labeled substrate in a buffer containing 0.5 mM EDTA, 75 mM sodium carbonate buffer (pH 9.2) at 90°C for 20 min. RNase T1 digestions were conducted by incubating the 30-mer 5′-end-labeled probe in 15 mM sodium citrate buffer (pH 5.0), 1 mM EDTA and 3.5 M Urea with 1 U of RNase T1 (Sigma) at 55°C for 15 min. All reactions were stopped with an equal volume of loading buffer containing 5 mM Tris, 5 mM boric acid, 0.05% bromophenol blue and 10 M Urea. The reaction products were analyzed on 17% denaturing polyacrylamide gel containing 8 M Urea and visualized by autoradiography.
Northern blotting analysis
The mRNA transcripts coding for the human RNase κ from various human tissues and organs were determined by northern blot analysis. Human MTN Blot hybridization membrane, containing 2 μg poly(A)+ RNA from eight different human tissues (heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas), was used. Hybridization of the 32P-labelled cDNA probe corresponding to nucleotides 1–466 of the human RNase κ cDNA sequence was performed at 68°C overnight in 25 ml ExpressHybTM Hybridization Solution (Clontech). Following hybridization the membrane was rinsed several times in 2× SSC, 0.05% SDS at room temperature, twice for 20 min in 0.5× SSC, 0.1% SDS at 50°C and twice for 10 min in 0.1× SSC, 0.1% SDS at 50°C. The blot was then covered immediately with plastic wrap, mounted on Whatman paper and exposed to X-ray film (Fuji film) at –80°C with two intensifying screens.
Identification of RNase κ family members
The dbEST (Expessed Sequence Tags) (26) database on the National Center for Biotechnology Information World Wide Web server was searched by the TBLASTN software (27). Multiple sequence alignment was performed by the Clustal W 1.8 program (28) using default parameters.
RESULTS
Identification and isolation of the human RNase κ cDNA
In previous work conducted in our laboratory, we presented the isolation and characterization of a novel ribonuclease, designated as Cc RNase, which bears high similarity with a group of sequences of unknown function, expanding from Caenorhabditis elegans to humans (19). A TBLASTN search of the available human EST sequence databases, using the deduced amino acid sequence of the homologous Cc RNase as query, resulted in the retrieval of a large number of EST sequences distributed in a variety of tissues. Based on the alignment of these sequences a set of specific oligonucleotide primers (FH and RH) was designed. In order to isolate the human RNase κ cDNA, 1 μg of poly(A)+ RNA from human placenta was reverse transcribed and the retrieved single-strand cDNA molecules were amplified by PCR using the FH and RH primers. The purified amplification product was cloned into the PCR™ 2.1 vector and five cDNA clones were selected and subjected to sequence analysis. Sequencing results (EMBL database, Accession No. AM746459) revealed no differences among these clones, which additionally appear to be identical with three of the previously submitted in the UniGene NCBI database (Hs.632232) mRNA sequences (Acc No. BC051802.1, Acc No. NM001004333.2 and Acc No. BC062705.1). The isolated cDNA clone has a length of 466 bp and includes an ORF of 297 nt encoding a protein of 98 amino acids with a calculated molecular mass of 10 984 kDa.
The gene encoding the human RNase κ (MGC71993) is located on chromosome 17, map position 17p13.1, corresponding to the LOC440400 gene locus. Genomic DNA and cDNA sequencing comparisons revealed that the human RNase κ gene consists of three exons interrupted by two introns with a length of 904 and 402 bp, respectively. The first exon (118 bp) includes 40 bp of the 5′ untranslated region and encodes the first 26 amino acids of the human protein. The second exon has a length of 77 bp, which code for the next 26 amino acids, and the third one consists of 387 bp, 139 bp of which encode the last 46 amino acids, while the rest 245 nucleotides belong to the 3′ untranslated region.
Conservation of the RNase κ family
Using the default settings of NCBI and the deduced amino acid sequence of the human RNase κ as query, a TBLASTN search of the available EST sequence databases was conducted. This search resulted in the retrieval of a large number of EST sequences bearing high similarity to human RNase κ, which expand throughout the animal kingdom. Multiple alignments of the deduced amino acid sequences belonging to different mammalian representatives indicated that the human RNase κ is an extremely conserved protein. It displays 100% identity compared to the predicted homologous proteins of monkeys, mice, rats and sheep, while cattle and pig proteins bear only one and two amino acid substitutions, respectively (Figure 1).
Figure 1.
Amino acid alignment of the human RNase κ and Cc RNase with the predicted amino acid sequences retrieved from ESTs belonging to other mammals. M. mulatta (Rhesus monkey): CB549243; M. musculus (mouse): BC021603; R. norvegicus (rat): BP467900; O. aries (sheep): EE797850; B. taurus (cattle): EH376754; S. scrofa (pig): DB782459) and C. capitata AJ441124. Identical amino acids are presented as dots and gaps as dashes.
Expression analysis of the human RNase κ mRNA
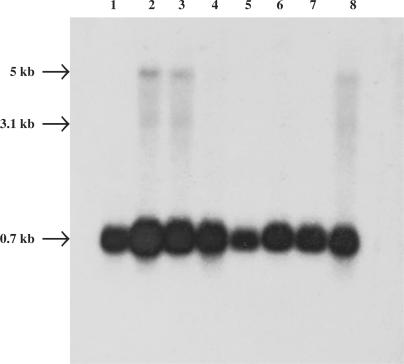
The TBLASTN search of the available human EST sequence databases resulted in the retrieval of a large number of EST clones (412 up today) prepared from a variety of normal (placenta, brain, kidney, etc.) and cancer tissues (osteosarcoma, breast adenocarcinoma, insulinoma, retinoblastoma, etc.). In order to confirm the organ distribution of the human RNase κ, a human multiple tissue northern blot membrane was hybridized using the 466 bp human RNase κ cDNA probe, as described in the Materials and Methods section. As shown in Figure 2, northern hybridization displayed the existence of a main mRNA transcript with an estimated length of 700 nt expressed in all tissues examined. This length is in agreement with that of the submitted BC051802.1, NM001004333.2 and BC062705.1 mRNA sequences mentioned above. It is interesting to point out that two additional less abundant mRNA transcripts of 3.1 and 5 kb, respectively, were identified only in brain, placenta and pancreas.
Figure 2.
Tissue distribution of the human RNase κ mRNA. PolyA + RNA (2 μg) on a human multiple tissue blot (Clontech) were hybridized to a 32P-labeled cDNA probe corresponding to nucleotides 1–466 of the human RNase κ cDNA sequence. 1: heart, 2: brain, 3: placenta, 4: lung, 5: liver, 6: skeletal muscle, 7: kidney, 8: pancreas.
Concerning the distribution of EST sequences among normal and cancer human tissues the vast majority of the ESTs (almost 75%) is represented in cancer tissues and only 25% in normal tissues. The wide expression of human RNase κ in multiple human adult tissues as well as in fetal, juvenile tissues and carcinomas suggests a basic housekeeping cellular function for this protein.
Expression of the human RNase κ in E. coli
The coding region of the human RNase κ cDNA was cloned into a variety of prokaryotic expression vectors including pET 15b, pET 20b, pET 29b, pET 39b, pET coco-1 and pSCREEN-1b(+). Most of our efforts to express the human enzyme using these vectors and a wide variety of E. coli host cells [Tunner(DE3), BL21(DE3), BL21(DE3)plysS, Rosetta(DE3), Rosetta(DE3)plysS] resulted either in lack of transformants or in severe toxic effects following IPTG induction. The inhibitory effect was not eliminated even when host cells harboring the plysS plasmid, which produces lysozyme as an additional means of reducing the recombinant protein expression level, were used.
An alternative expression system, in which T7 RNA polymerase is provided by infection with the bacteriophage CE6 (29) and is widely used for the expression of extremely toxic genes, was also tested. Despite the fact that this system has previously been successfully used for the expression of the toxic ribonuclease RegB (30), efforts to express the human RNase κ resulted again in severe cell death.
Successful expression was achieved only after transformation of the recombinant pSCREEN-1b(+) vector into BL21(DE3)plysS host cells. In this construct the target protein is expressed as a fusion protein containing the first 260 amino acids of the bacteriophage T7 gene 10 major capsid protein at its N-terminus. Although the use of this expression system resulted in the production of large amounts of insoluble fusion protein, the mature purified recombinant protein exhibited very low ribonucleolytic activity against synthetic homopolyribonucleotides (data not shown).
Expression of the human RNase κ in P. pastoris
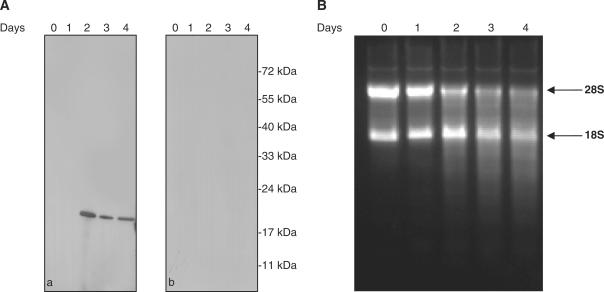
In order to overcome the problems related to the expression of the human RNase κ in prokaryotic systems, the methylotrophic yeast P. pastoris expression system was employed. The human RNase κ cDNA was subcloned into pPICZα A expression vector, creating the pαA construct. In this construct the target protein is expressed as a small fusion protein containing a c-myc and a His-tag epitope at its C-terminus. Furthermore, a signal peptide facilitates the secretion of the produced recombinant protein in the culture medium. The obtained transformants were selected on the basis of high antibiotic resistance and the integration of the human RNase κ cDNA into the genomic yeast DNA was confirmed by PCR using the 5′ AOX1 and 3′ AOX1 universal primers. The expression of the recombinant protein was verified by western blot analysis using the monoclonal anti His-tag antibody. As shown in Figure 3A, only one band with an approximate molecular mass of 19 kDa was detected 48 h after methanol induction in the culture medium, while no signal was observed in a parallel control experiment. The cultures’ supernatants were also tested for ribonucleolytic activity against rRNA. As shown in Figure 3B, the appearance of ribonucleolytic activity in the cultures’ supernatants follows the expression and secretion of the recombinant protein.
Figure 3.
Detection of the recombinant protein secreted in P. pastoris culture supernatant by immunoblotting analysis (A) and ribonucleolytic activity (B). (A) Equal aliquots of the culture supernatant collected at 0, 1, 2, 3 and 4 days after methanol induction were analyzed by SDS–PAGE, electro-transferred onto a nitrocellulose membrane and the expressed recombinant protein was detected using a monoclonal antibody against the His-tag epitope and a goat anti-mouse IgG antibody conjugated to horseradish peroxidase (a). In a parallel experiment using only the second antibody no bands were detected (b). (B) An aliquot of 2 μg of rRNA isolated from rat liver was incubated at 37°C for 30 min with 5 μl of supernatant from the Pichia pastoris cultures collected at 0, 1, 2, 3 and 4 days after induction. The reaction products were analyzed by 1% agarose gel electrophoresis and visualized with ethidium bromide staining. Arrows indicate 28S and 18S rat rRNA.
Purification and characterization of the expressed human RNase κ
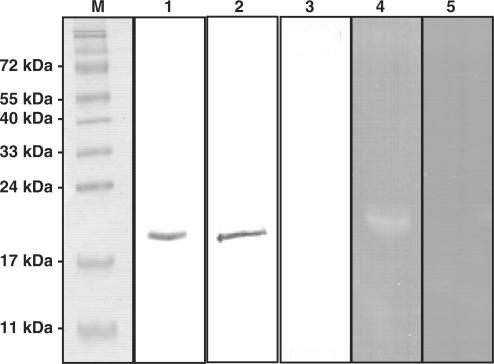
The human RNase κ secreted in the culture medium was purified under native conditions using the Protino His-Bind Resin as described in Materials and Methods section. The purified protein appeared as a single band and exhibited a molecular mass of ∼19 kDa as estimated by SDS–PAGE (Figure 4, lane 1). The identity of the purified protein was immunologically verified using the polyclonal antibody raised in chicken against a specific oligopeptide of the human RNase κ, as described in the Materials and Methods section. Using this antibody a single band migrating at the same position on the SDS–PAGE was detected (Figure 4, lane 2). No immunodetection was obtained when the same sample was treated only with the second antibody (Figure 4, lane 3). When the ribonucleolytic activity of the purified fraction was checked by SDS–PAGE in the presence of poly (U), a single band corresponding to the molecular mass of the purified protein was detected in the gel followed by zymography (Figure 4, lane 4). In order to prove that this band could not be attributed to contaminating ribonucleolytic activity, it is significant to mention that no band appeared in the activity gel when the purified recombinant protein was preincubated at 99°C for 5 min, conditions which inactivate the human RNase κ (Figure 4, lane 5).
Figure 4.
SDS–PAGE, immunoblot analysis and activity staining of the purified recombinant human RNase κ. An aliquot of 2 μg of the purified human RNase κ was analyzed by SDS–PAGE and stained with Coomassie blue (lane 1). A set of marker proteins of known molecular weight (72, 55, 40, 33, 24, 17 and 11 kDa top to bottom) were run in parallel (lane M). Aliquots of the same amount of the purified recombinant protein were analyzed by SDS–PAGE, electro transferred onto a nitrocellulose membrane and detected using a polyclonal antibody raised against a specific chemically synthesized amino acid sequence of the human RNase κ and a goat anti-chicken antibody conjugated to alkaline phosphatase (lane 2). In a parallel experiment using only the second antibody no band was detected (lane 3). An aliquot of 2 μg of the purified recombinant protein (lane 4) and an equivalent amount of this protein preincubated at 99°C for 5 min (lane 5) were analyzed on a 15% polyacrylamide gel containing 0.25 mg/ml poly (U). After electrophoresis the gel was stained for RNase activity as described in Materials and Methods section.
It is also important to note that the calculated mass deduced from the amino acid sequence of the human RNase κ expressed by the pαA construct in P. pastoris (13 937 kDa) is deviated from that estimated by SDS–PAGE. Therefore, in order to confirm further the identification of the purified recombinant protein, the pure elution fragment was analyzed on SDS–PAGE and the 19 kDa band was excised, digested with trypsin and the resulted peptides were extracted and subjected to LC-ESI-MS and MALDI-MS analysis. Data obtained from this procedure resulted in the identification of a number of distinct peptide fragments covering 80% of the human RNase κ amino acid sequence.
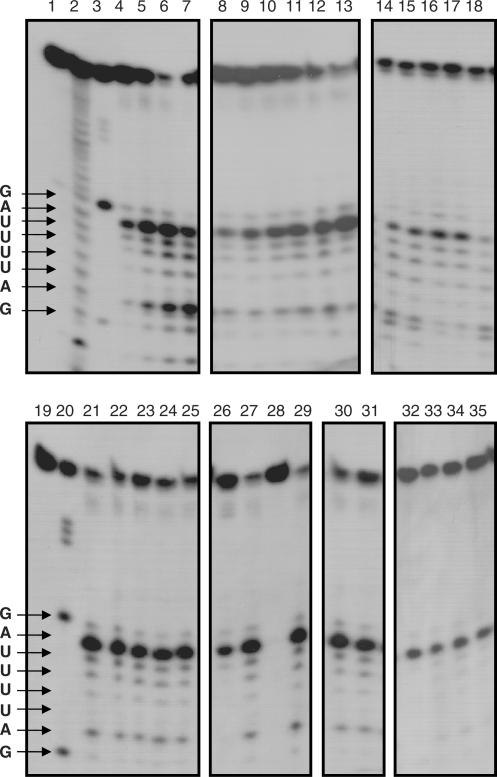
The substrate specificity of the recombinant human RNase κ against poly (U), poly (C) and torula RNA was determined by evaluating the generation of acid-soluble ribonucleotides, as described in Materials and Methods section. The recombinant enzyme degrades preferentially poly (U), while it degrades torula yeast RNA at a lower rate. No degradation of poly (C) was detected under the same experimental conditions, although some minor degradation of this substrate was observed after extended incubation times and increased enzyme concentrations. The human RNase κ base specificity was determined by incubation of the purified recombinant protein with a 5′-32P-labeled synthetic 30-mer RNA with known sequence, followed by denaturing gel electrophoresis and autoradiography. As shown in Figure 5, lanes 4–7, the ApU phosphodiester bond is mainly cleaved at the lowest enzyme concentration, while increase of the enzyme concentration enhances the cleavage of the ApG and UpU phosphodiester bonds. The cleavage efficiency of the preferred phosphodiester bonds is in correlation with increasing incubation time (Figure 5, lanes 8–13).
Figure 5.
Base specificity and enzymatic characterization of human RNase κ. A 30-mer 5′-labeled oligoribonucleotide of known sequence was incubated without RNase κ (lanes 1 and 19), in alkaline hydrolysis buffer (lane 2) or with 1 U of ribonuclease T1 (lanes 3 and 20). The same amount of substrate was incubated at 37°C with different amounts of purified recombinant protein (20, 50, 100, 200 ng) for 30 min (lanes 4–7, respectively) or with 30 ng of purified recombinant protein for 10, 20, 30, 40, 50 and 60 min (lanes 8–13, respectively). For the determination of the optimum pH of the human RNase κ, the 5′end-labeled probe was incubated with 25 ng of recombinant protein in 50 mM acetate buffer pH 5.0 and 6.0 (lanes 14 and 15) or 50 mM Tris–HCl pH 7.0, 8.0 and 9.0 (lanes 16–18). For the enzymatic characterization of the human RNase κ, the 5'end labelled probe was incubated with 25 ng of recombinant protein (except indicated otherwise) in 20 mM Tris–HCl pH 8.0 at 37°C for 30 min (lane 21); or in the presence of 5 mM EDTA (lane 22); 100, 150 and 250 mM NaCl (lanes 23–25); 2 mM Cu+2, Mg+2, Zn+2 and Mn+2 (lanes 26–29, respectively); 50 mM NEM or iodoacetamide (lanes 30 and 31, respectively). Lanes 32–35: 10 ng human RNase κ with 0, 10, 20 and 40 U human placental inhibitor (RNasin). The partially digested RNA probe was separated by 8 M urea–17% polyacrylamide gel electrophoresis and visualized by autoradiography.
The biochemical properties of the studied human RNase κ were tested using the 30-mer 5′-end-labeled RNA probe as substrate. The enzyme is active in a wide pH range from 5 to 8 (Figure 5, lanes 14–18). The sequence base specificity of this RNase is not affected by the presence of EDTA up to 20 mM (Figure 5, lane 22) or NaCl up to 250 mM (Figure 5, lanes 23–25). Addition of the alkylating agent NEM or iodoacetamide does not inhibit the RNase activity of the recombinant enzyme in concentrations up to 50 mM (Figure 5, lanes 30 and 31). The purified enzyme seems to remain unaffected in the presence of Mg+2 and Mn+2, while 2 mM Cu+2 or Zn+2 cause slight and total inhibition of its ribonucleolytic activity, respectively (Figure 5, lanes 26–29). Addition of the reducing agent DTT in concentrations > 5 mM causes its complete inactivation leading to the conclusion that the formation of disulfide bonds is essential for the retention of enzymatic activity. On the other hand, the presence of the placental ribonuclease inhibitor has no effect on the activity of the human RNase κ, even at very high amounts (40 U) and limiting enzyme concentration (Figure 5, lanes 32–35). This finding adds an additional argument to the fact that the RNase κ family differs significantly from the well-characterized RNase A superfamily. The effect of temperature on ribonucleolytic activity was determined after preincubation of the recombinant enzyme at 37, 60 and 90°C for various time periods (data not shown). This experiment proved that the human RNase κ is a thermolabile molecule, since it loses most of its activity after preincubation at 60°C for 10 min.
DISCUSSION
Ribonucleases can be considered as more than simple RNA digesting enzymes. They act as messenger molecules by interacting with actin, heparin and proteoglycans and influence a wide range of functions. Their degrading action on mRNA molecules has been under scrutiny for a number of years, as this procedure seems to play an important role in gene expression or translational control and therefore in cell differentiation. Besides their digestive role many RNases exhibit anti-tumor and cytotoxic activity, while others exert aspermatogenic, anti-embryonic, anti-bacterial, anti-viral or immunosuppressive actions. In our previous work, we presented a novel RNase, Cc RNase, isolated from the insect Ceratitis capitata, which belongs to a novel highly conserved protein family (19).
In this work, we report the isolation and characterization of a cDNA clone encoding the human counterpart of the RNase κ (kappa) family along with the expression and biochemical characterization of the recombinant enzyme. Search of EST sequences databases, belonging to a variety of different organisms, proved that the RNase κ family is represented by a single ortholog in metazoans expanding from C. elegans to humans, while no representatives have been detected so far in yeast, plants and bacteria. All members of this family are small proteins with polypeptide chains consisting of 95–101 amino acids and have no significant similarity to other known ribonucleases. It is important to note that all members of this family seem to be highly conserved with a conservation rate that in mammals reaches a percentage > 98%. This extremely high conservation can be compared only to the increased conservation rates exhibited by members of other protein families with very significant biological roles, such as the histones (31) or phosphofructokinase (32).
As far as the genomic organization of the human RNase κ is concerned, the human RNase κ gene is represented by a single copy in the human genome and retains the pattern of gene organization that is preserved by all known members of the gene family, consisting of three exons interrupted by two introns.
Northern blot analysis resulted in the detection of a main mRNA of the human RNase κ exhibiting a length of 700 kb in all tissues examined. Two longer transcripts of 3.1 and 5 kb were found only in brain, placenta and pancreas, while a third one with a length of 1.6 kb expressed in B cells (Ramos cell line) has already been submitted in the databank (Acc No. CR607998.1). Analysis of the retrieved EST sequences revealed that the human RNase κ mRNA is expressed in almost every human tissue and developmental stage. This finding suggests a basic housekeeping cellular function for this protein. Concerning the breakdown of EST sequences of the human RNase κ by health state, ∼75% of them are represented in cancer tissues while the rest in normal tissues.
In most cases, the expression of the human RNase κ in bacteria caused a severe toxic effect, resulting either in the production of no transformants or in the inhibition of cell growth and death of the transformed E. coli cultures following IPTG induction. It is already known that the expression of many human recombinant RNases with important biological actions, such as ECP (23) or human RNase 7 (33) produces similar toxic effects. The toxic effect produced by the expression of the human RNase κ was not eliminated even when host cells, which produce lysozyme as an additional means of reducing basal expression of the recombinant protein or the expression system of the bacteriophage CE6 were used. It is therefore possible that the production of even a small amount of the recombinant protein is sufficient to cause a severe toxic effect in E. coli. The successful expression of the human RNase κ in bacteria was achieved only after the recombinant molecule was expressed as a fusion protein. While this expression system produced a large amount of insoluble fusion protein, an extremely low amount of soluble human RNase κ, which exhibited very little ribonucleolytic activity, yielded after purification.
Therefore, in order to express the human RNase κ, the methylotrophic yeast P. pastoris expression system, which has been previously used successfully for the expression of other toxic ribonucleases, such as the a-sarcin (34) or onconase (35), was employed. Using this system, the production of a highly active recombinant ribonucleolytic enzyme was accomplished. Even though the calculated molecular weight of the secreted recombinant protein is almost 14 kDa, the purified protein displays a molecular mass of 19 kDa as estimated by SDS–PAGE. Despite this deviation the identity of the recombinant protein is unquestionable, since the 19 kDa protein exhibits ribonucleolytic activity, immunoreacts with the polyclonal antibody raised against a specific oligopeptide of the human RNase κ as well as with the monoclonal anti-His tag antibody and yields the expected peptide mapping profile as resulted by ESI-MS and MALDI-MS analysis. Since the results obtained from the peptide analysis did not report identification of modified amino acids, the observed difference among the theoretical and experimental molecular weights could probably be attributed to an abnormal electrophoretic mobility of the recombinant protein, as has been previously reported in the case of the recombinant ribonuclease U2 (36).
As far as the catalytic mechanism employed by the human RNase κ in the degradation of RNA is concerned, the existence of one histidine residue (His36) in its amino acid sequence raises a significant point as to whether this residue can take part in the catalytic mechanism. Two histidine residues participate in acid–base catalysis caused by all members of the RNase A superfamily (13), whereas one histidine and one acidic amino acid are necessary for the degradation of RNA from the fungal RNase T1 (37). On the other hand, there are also ribonucleases such as RNase III and RNase H type II, by which RNA catalysis is carried out by acidic amino acid residues only (38–39). Although experimental data concerning the catalytic mechanism of the human RNase κ have not been obtained so far, residues Lys10, Asp43, Tyr62, Tyr76, Gln86 and Asn90 of the human enzyme are found to be conserved in all family members studied, therefore indicating a possible participation in catalysis or in RNA base-specific recognition.
The human RNase κ contains five cysteine residues, three of which Cys7, Cys14 and Cys69 are conserved among all species examined. The finding that the recombinant enzyme loses its activity by the addition of DTT in the reaction mixture, while it remains fully active in the presence of N-ethylmaleimide or iodoacetamide leads us to the conclusion that cysteine residues are not necessary for RNA catalysis, but stabilize the protein by the formation of at least one disulfide bond.
Base specificity experiments indicated that the human RNase κ is probably a single-strand-specific endoribonuclease that preferentially cleaves ApU and ApG phosphodiester bonds, while it hydrolyzes UpU bonds at a lower rate. This novel RNase has no obvious relationship with the pancreatic RNase family enzymes and seems more similar to other human RNases, like RNase T84 (40), RNase L (41) or RNase 4 (42). However, these enzymes recognize different dinucleotides than the human RNase κ. For example, RNase T84 catalyzes the hydrolysis of ApU and GpU phosphodiester bonds, RNase L catalyzes the cleavage of UpU, UpA, UpG, ApA and ApU, while RNase 4 hydrolyzes the phosphodiester bonds of UpA and UpG.
Human RNase κ sequence specificity also differs from the specificity for G and A nucleotides known for fungal RNases, RNase T1 (43) and RNase U2 (44), respectively. Furthermore, it seems to differ from the fungal RNase Phy M (45), which cleaves RNA after U and A nucleotides. The latter enzymes recognize all guanosine, adenine or uracil nucleotides in the single-stranded RNA regions and do not require a following nucleotide.
A comparison of the human RNase κ with its previously characterized C. capitata counterpart has displayed a number of significant similarities between them. The human protein is three amino acids longer consisting of 98 amino acids and displays 40% identity in the amino acid level with Cc RNase. Most of our efforts to express these proteins in prokaryotic systems resulted either in lack of transformants or severe cell death following IPTG induction, thus indicating that the production of either of these RNases is toxic to E. coli. In both cases successful expression was achieved only by the use of the pSCREEN 1b(+) vector, by which the target RNases are expressed as fusion proteins. Even though the use of this system produced satisfactory amounts of recombinant fusion proteins, ribonucleolytically active mature protein yielded only in the case of Cc RNase. The biochemical characterization of the human and insect protein family representatives has proved that both enzymes hydrolyze native and synthetic RNAs at the same pH range, are thermolabile and are inhibited by Zn+2. Reduction of their disulfide bonds results in their complete inactivation indicating that the formation of at least one disulfide bond is essential for their catalytic activity. Concerning the nucleotide base specificity both enzymes cleave UpU phosphodiester bonds, while Cc RNase cleaves CpC bonds more effectively compared to its human counterpart. Even though the ribonucleolytic activity of Cc RNase has been characterized only by the use of homopolymers, the finding that the human RNase κ displays a deviated specificity is not unusual since similar data have been also reported in the case of other RNase families. For example, frog ribonucleases of the RNase A superfamily display a preference for the hydrolysis of pyrimidine-G phosphodiester bonds compared to the preference for pyrimidine-A phosphodiester bonds displayed by their mammalian counterparts (46). Furthermore, it is known that the specificity of dinucleotide cleavage by angiogenins differs among distinct family members (47). Additionally, the RNase T1 variant RV, bearing only five amino acid substitutions, exhibits a stronger preference toward adenosine residues than wild-type RNase T1 (48).
In summary, the characterization of the human RNase κ as the second member of a novel RNase family establishes the identification of the RNase κ family in metazoans. The nucleotide base specificity of the recombinant human representative, its lack of any significant homology to the members of the RNase A family and the fact that the human counterpart evades the placental ribonuclease inhibitor prove that there is no evolutionary relationship between RNase A and κ families. The high conservation of all members of this RNase family in combination with the fact that the human enzyme is found to be expressed in all developmental stages and tissues suggest a very important biological function, which remains to be elucidated. Additionally, since it is well known that a number of human RNases are involved with pathogenic conditions, like autoimmune diseases, inflammatory disorders (49) or inhibition of tumor growth and metastasis (50), a possible participation of the human RNase κ in the occurrence of such pathological situations would be intriguing to be explored.
ACKNOWLEDGEMENTS
This work is part of Ph.D. thesis of Ms E.A.M. and was supported by the Hellenic State Scholarship's Foundation and a Faculty Research Grant from the National Kapodistrian University of Athens. Funding to pay the Open Access publication charges for this article was provided by the University of Athens.
Conflict of interest statement. None declared.
REFERENCES
- 1.D’ Alessio G, Riordan JF. Ribonucleases, Structures and Functions. San Diego, CA, USA: Academic Press; 1997. [Google Scholar]
- 2.Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 3.Fett JW, Strydom DJ, Lobb RR, Alderman EM, Bethune JL, Riordan JF, Vallee BL. Isolation and characterization of angiogenin, an angiogenic protein from human carcinoma cells. Biochemistry. 1985;24:5480–5486. doi: 10.1021/bi00341a030. [DOI] [PubMed] [Google Scholar]
- 4.Rosenberg HF, Tenen DG, Ackerman SJ. Molecular cloning of the human eosinophil-derived neurotoxin: a member of the ribonuclease gene family. Proc. Natl Acad. Sci. USA. 1989;86:4460–4464. doi: 10.1073/pnas.86.12.4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosenberg HF, Ackerman SJ, Tenen DG. Human eosinophil cationic protein. Molecular cloning of a cytotoxin and helminthotoxin with ribonuclease activity. J. Exp. Med. 1989;170:163–176. doi: 10.1084/jem.170.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rosenberg HF, Dyer KD. Human ribonuclease 4 (RNase 4): coding sequence, chromosomal localization and identification of two distinct transcripts in human somatic tissues. Nucleic Acids Res. 1995;23:4290–4295. doi: 10.1093/nar/23.21.4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenberg HF, Dyer KD. Molecular cloning and characterization of a novel human ribonuclease (RNase k6): increasing diversity in the enlarging ribonuclease gene family. Nucleic Acids Res. 1996;24:3507–3513. doi: 10.1093/nar/24.18.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Larson KA, Olson EV, Madden BJ, Gleich GJ, Lee NA, Lee JJ. Two highly homologous ribonuclease genes expressed in mouse eosinophils identify a larger subgroup of the mammalian ribonuclease superfamily. Proc. Natl Acad. Sci. USA. 1996;93:12370–12375. doi: 10.1073/pnas.93.22.12370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beintema JJ, Kleineidam RG. The ribonuclease A superfamily: general discussion. Cell. Mol. Life Sci. 1998;54:825–832. doi: 10.1007/s000180050211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Dyer KD, Rosenberg HF. Evolution of the rodent eosinophil-associated RNase gene family by rapid gene sorting and positive selection. Proc. Natl Acad. Sci. USA. 2000;97:4701–4706. doi: 10.1073/pnas.080071397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liao YD, Huang HC, Leu YJ, Wei CW, Tang PC, Wang SC. Purification and cloning of cytotoxic ribonucleases from Rana catesbeiana (bullfrog) Nucleic Acids Res. 2000;28:4097–4104. doi: 10.1093/nar/28.21.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenberg HF, Zhang J, Liao YD, Dyer KD. Rapid diversification of RNase A superfamily ribonucleases from the bullfrog Rana catesbeiana. J. Mol. Evol. 2001;53:31–38. doi: 10.1007/s002390010188. [DOI] [PubMed] [Google Scholar]
- 13.Zhang J, Dyer KD, Rosenberg HF. RNase 8, a novel RNase A superfamily ribonuclease expressed uniquely in placenta. Nucleic Acids Res. 2002;30:1169–1175. doi: 10.1093/nar/30.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Zhang YP, Rosenberg HF. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nat. Genet. 2002;30:411–415. doi: 10.1038/ng852. [DOI] [PubMed] [Google Scholar]
- 15.Benner SA, Allemann RK. The return of pancreatic ribonucleases. Trends Biochem. Sci. 1989;14:396–397. doi: 10.1016/0968-0004(89)90282-x. [DOI] [PubMed] [Google Scholar]
- 16.D’Alessio G, Di Donato A, Parente A, Piccoli R. Seminal RNase: a unique member of the ribonuclease superfamily. Trends Biochem. Sci. 1991;16:104–106. doi: 10.1016/0968-0004(91)90042-t. [DOI] [PubMed] [Google Scholar]
- 17.Vallee BL, Riordan JF. Organogenesis and angiogenin. Cell Mol. Life Sci. 1997;53:803–815. doi: 10.1007/s000180050101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soochin C, Beintema JJ, Zhang J. The ribonuclease A superfamily of mammals and birds: identifying new members and tracing evolutionary histories. Genomics. 2005;85:208–220. doi: 10.1016/j.ygeno.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 19.Rampias TN, Sideris DC, Fragoulis EG. Cc RNase: the Ceratitis capitata ortholog of a novel highly conserved protein family in metazoans. Nucleic Acids Res. 2003;31:3092–3100. doi: 10.1093/nar/gkg414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 21.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl Acad. Sci. USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 23.Rosenberg HF. Recombinant human eosinophil cationic protein. Ribonuclease activity is not essential for cytotoxicity. J. Biol. Chem. 1995;270:7876–7881. doi: 10.1074/jbc.270.14.7876. [DOI] [PubMed] [Google Scholar]
- 24.Slifman NR, Loegering DA, McKean DJ, Gleich GJ. Ribonuclease activity associated with human eosinophil-derived neurotoxin and eosinophil cationic protein. J. Immunol. 1986;137:2913–2917. [PubMed] [Google Scholar]
- 25.Blank A, Sugiyama RH, Dekker CA. Activity staining of nucleolytic enzymes after sodium dodecyl sulfate-polyacrylamide gel electrophoresis: use of aqueous isopropanol to remove detergent from gels. Anal. Biochem. 1982;120:267–275. doi: 10.1016/0003-2697(82)90347-5. [DOI] [PubMed] [Google Scholar]
- 26.Boguski MS, Lowe TM, Tolstoshev CM. dbEST- database for expressed sequence tags. Nat. Genet. 1993;4:332–333. doi: 10.1038/ng0893-332. [DOI] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhahg Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Julie D, Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miao F, Drake SK, Kompala DS. Characterization of gene expression in recombinant Escherichia coli cells infected with phage lambda. Biotechnol. Prog. 1993;9:153–159. doi: 10.1021/bp00020a006. [DOI] [PubMed] [Google Scholar]
- 30.Saida F, Uzan M, Bontems F. The phage T4 restriction endoribonuclease RegB: a cyclizing enzyme that requires two histidines to be fully active. Nucleic Acids Res. 2003;31:2751–2758. doi: 10.1093/nar/gkg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pehrson RJ, Fuji RN. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 1998;26:2837–2842. doi: 10.1093/nar/26.12.2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kao MC, Wuu JA, Pai SH. Comparison of 5' region sequences of human and rabbit phosphofructokinase genes. Proc. Natl Sci. Counc. Repub. China. 1990;14:15–19. [PubMed] [Google Scholar]
- 33.Zhang J, Dyer KD, Rosenberg HF. Human RNase 7: a new cationic ribonuclease of the RNase A superfamily. Nucleic Acids Res. 2003;31:602–607. doi: 10.1093/nar/gkg157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martinez-Ruiz A, Martinez del Pozo A, Lacadena J, Mancheno JM, Onaderra M, Lopez-otin C, Gavilanes JG. Secretion of recombinant pro- and mature fungal alpha-sarcin ribotoxin by the methylotrophic yeast Pichia pastoris: the Lys-Arg motif is required for maturation. Protein Expr. Purif. 1998;12:315–322. doi: 10.1006/prep.1997.0846. [DOI] [PubMed] [Google Scholar]
- 35.Kim BM, Kim H, Raines RT, Lee Y. Glycosylation of onconase increases its conformational stability and toxicity for cancer cells. Biochem. Biophys. Res. Commun. 2004;315:976–983. doi: 10.1016/j.bbrc.2004.01.153. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Ortega L, De los Rios V, Martinez-Ruiz A, Onaderra M, Lacadena J, Martinez del Pozo. A, Gavilanes JG. Anomalous electrophoretic behaviour of a very acidic protein: ribonuclease U2. Electrophoresis. 2005;26:3407–3413. doi: 10.1002/elps.200500261. [DOI] [PubMed] [Google Scholar]
- 37.Steyaert J, Hallenga K, Wyns L, Stanssens P. Histidine-40 of ribonuclease T1 acts as base catalyst when the true catalytic base, glutamic acid-58, is replaced by alanine. Biochemistry. 1990;29:9064–9072. doi: 10.1021/bi00490a025. [DOI] [PubMed] [Google Scholar]
- 38.Sun W, Nicholson AW. Mechanism of action of Escherichia coli ribonuclease III. Stringent chemical requirement for the glutamic acid 117 side chain and Mn2+ rescue of the Glu117Asp mutant. Biochemistry. 2001;40:5102–5110. doi: 10.1021/bi010022d. [DOI] [PubMed] [Google Scholar]
- 39.Chapados BR, Chai Q, Hosfeld DJ, Qiu J, Shen B, Tainer JA. Structural biochemistry of a type 2 RNase H: RNA primer recognition and removal during DNA replication. J. Mol. Biol. 2001;307:541–556. doi: 10.1006/jmbi.2001.4494. [DOI] [PubMed] [Google Scholar]
- 40.Przewlocki G, Lipecka J, Edelman A, Przykorska A. New sequence-specific human ribonuclease: purification and properties. Nucleic Acids Res. 1998;26:4047–4055. doi: 10.1093/nar/26.17.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Floyd-Smith G, Slattery E, Lengyel P. Interferon action: RNA cleavage pattern of a (2′-5′) oligoadenylate-dependent endonuclease. Science. 1981;212:1030–1032. doi: 10.1126/science.6165080. [DOI] [PubMed] [Google Scholar]
- 42.Shapiro R, Fett JW, Strydom DJ, Vallee BL. Isolation and characterization of a human colon carcinoma-secreted enzyme with pancreatic ribonuclease-like activity. Biochemistry. 1986;25:7255–7264. doi: 10.1021/bi00371a002. [DOI] [PubMed] [Google Scholar]
- 43.Irie M. Enzymatic depolymerization of synthetic polynucleotides, poly A, poly C and poly U by ribonuclease T1 preparations. J. Biochem. 1965;58:599–603. doi: 10.1093/oxfordjournals.jbchem.a128249. [DOI] [PubMed] [Google Scholar]
- 44.Uchida T, Arima T, Egami F. Specificity of RNase U2. J. Biochem. 1970;67:91–102. doi: 10.1093/oxfordjournals.jbchem.a129239. [DOI] [PubMed] [Google Scholar]
- 45.Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980;8:3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liao YD, Huang HC, Leu YJ, Wei CW, Tang PC, Wang SC. Purification and cloning of cytotoxic ribonucleases from Rana catesbeiana (bullfrog) Nucleic Acids Res. 2000;28:4097–4104. doi: 10.1093/nar/28.21.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strydom DJ. The angiogenins. Cell Mol. Life Sci. 1998;54:811–824. doi: 10.1007/s000180050210. Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Czaja R, Struhalla M, Höschler K, Saenger W, Sträter N, Hahn U. RNase T1 variant RV cleaves single-stranded RNA after purines due to specific recognition by the Asn46 side chain amide. Biochemistry. 2004;43:2854–2862. doi: 10.1021/bi035961f. [DOI] [PubMed] [Google Scholar]
- 49.Schwartz B, Shoseyov O, Melnikova VO, McCarty M, Leslie M, Roi. L, Smirnoff P, Hu GF, Lev D, Bar-Eli M. ACTIBIND, a T2 RNase, competes with angiogenin and inhibits human melanoma growth, angiogenesis and metastasis. Cancer Res. 2007;67:5258–5266. doi: 10.1158/0008-5472.CAN-07-0129. [DOI] [PubMed] [Google Scholar]
- 50.Ohta N, Okazaki S, Fukase S, Akatsuka N, Aoyagi M, Yamakawa M. Serum concentrations of eosinophil cationic protein and eosinophils of patients with Kimura's disease. Allergol. Int. 2007;56:45–49. doi: 10.2332/allergolint.O-06-442. [DOI] [PubMed] [Google Scholar]