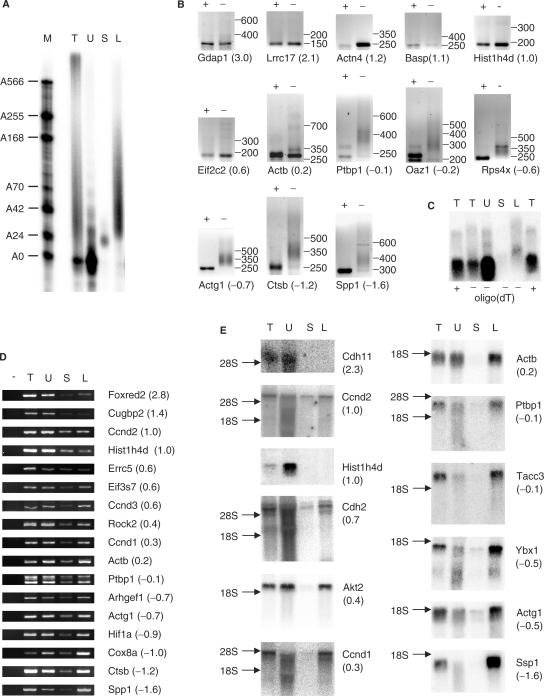
Figure 4.
Validation of poly(A) fractionation microarray. T, total RNA; U, unbound; S, short (oligoadenylated RNA eluted with 0.075× SSC); L, long (polyadenylated RNA eluted with H2O). Gene names are according to Genbank, numbers in brackets indicate the corrected LogRatio obtained for the mRNA in the microarray screen. A high LogRatio should correlate with a short poly(A) tail. (A) Total RNA from NIH3T3 cells was mixed with radiolabelled polyadenylated probe and biotinylated oligo(dT) and then fractionated using the poly(A) fractionation method. Ten percent of the resulting fractions was analysed by urea–PAGE as in Figure 3A. The lane with total RNA contains 2% of starting material. The remainder of the fractions with short and long poly(A) tails was used for microarray analysis (see Supplementary Table 2). (B) Total RNA from NIH3T3 was subjected to RNaseH treatment in the presence of oligo(dT) (+ lanes) or used untreated (− lanes) and subjected to RL-PAT using specific sense primers (Supplementary Table 1) for genes identified in the microarray analysis. The size difference in PCR products between RNaseH treated and untreated corresponds to the length of the poly(A) tail. The numbers after the gene name indicate the corrected LogRatio obtained in the microarray experiment. (C) Total RNA from NIH3T3 cells was treated with RNaseH in the presence of oligo(dT) and an antisense oligo specific for Actb (+) or the presence of the specific oligo only (−). The sample treated with the specific oligo only was subsequently fractionated using the poly(A) fractionation method (U, S, L). Actb RNA was detected by Northern analysis. (D) RNA was fractionated as described in (A). RT-PCR was performed on the resulting fractions. Samples in the no RT lane (−) were treated the same as samples in the total RNA lane (T) except for the omission of Superscript III. (E) RNA was fractionated as described in (A). Twenty-five percent of each fraction was then analysed by northern blotting. Total: 5% of starting material. The numbers in brackets behind the gene names refer to the average corrected LogRatio (see Supplementary Table 2).