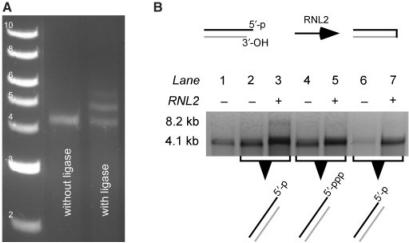
Figure 6.
Ligation of double-stranded RNA molecules with complementary 5′-overhangs using RNA Ligase 2 (RNL2). (A) Intermolecular ligation of a 0.5 kb dsRNA molecule to each end of a 4.1 kb dsRNA with RNL2 through complementary 5′-overhangs (Figure 2D) (gel image from (50), and used with the permission of NAR). Lanes: 1, double-stranded DNA marker. The corresponding dsDNA lengths are noted next to each band; 2, 4.1 kb dsRNA with 5′-overhang at both ends incubated in the RNL2 ligation buffer; 3, ligation of 0.5 kb dsRNA to the 3′- and 5′-ends of 4.1 kb dsRNA with RNL2. (B) The efficiency of intramolecular ligation with RNL2 was tested with a 4.1 kb dsRNA with a monophosphate at its 5′-overhang (activated dsRNA). Two control molecules 4.1 kb dsRNA with a triphosphate at its 5′-overhang (deactivated dsRNA), and the 4.1 kb dsRNA with a blunt end (blunt dsRNA) were also incubated with RNL2. Lanes: 1, 4.1 kb ssRNA; 2, activated dsRNA; 3, activated dsRNA ligated with RNL2; 4, deactivated dsRNA; 5, deactivated dsRNA ligated with RNL2; 6, blunt dsRNA; 7, blunt dsRNA ligated with RNL2. The unligated dsRNA molecules run as 4.1 kb ssRNA after denaturation whereas intramolecularly ligated dsRNA molecules run as 8.2 kb ssRNA after dentauration. The 4.1 and 8.2 kb ssRNA mobilities are indicated in the gel image.