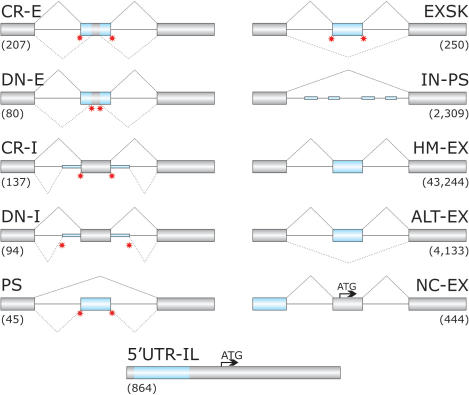
Figure 1.
Schematic representation of sequence categories. Primary transcripts are shown as exons (boxes) and introns (lines). Exonic or intronic sequences analysed in this study are shown as blue boxes or blue thick lines, respectively. Canonical and aberrant splicing events are shown above and below primary transcripts, respectively. Disease-associated splicing mutations are schematically shown as red stars. Designation of each sequence category and corresponding numbers of analysed sequences is above and below the primary transcript, respectively. Arrows denote translation initiation sites. CR-E, sequences between cryptic splice sites in exons and their authentic counterparts; DN-E, sequences between de novo splice sites in exons and their authentic counterparts; CR-I, cryptic splice sites in introns; DN-I, de novo splice sites in introns; PS, sequences of mutation-induced pseudoexons (Table S2); EXSK, sequences of exons that were skipped as a result of splice-site mutations leading to disease phenotypes (Table S1); IN-PS, intronic sequences that have strong 3′ss and 5′ss and a size of 50–250 nt, but were never used by the spliceosome (14); HM-EX, human exons homologous to mouse exons (30); ALT-EX, alternatively spliced human exons (30); NC-EX, non-coding exons lacking protein-coding information (14); 5′-UTR-IL, sequences of 5′UTR in intron-less genes (14). The total length of the sequence categories (in nucleotides, nt) was 10 383; 7862; 10 686; 6988; 6114; 29 292; 352 688; 5 817 754; 513 162; 50 187 and 245 076, respectively.