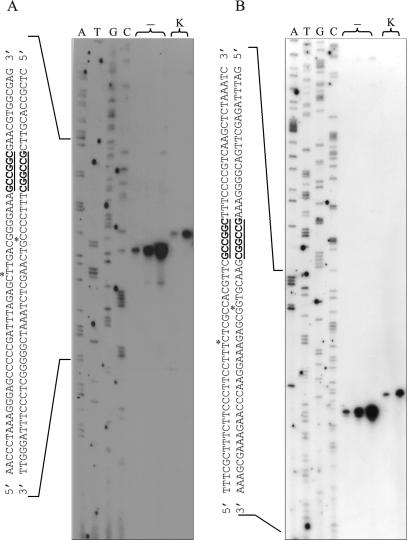
Figure 3.
Determination of the cleavage site of R.NmeDI. DNA sequencing reactions were performed on pBluescript KS II (+) DNA containing the R.NmeDI recognition site using pKSRDIf or pKSRDIr primers. Labeled test DNA was generated using the same primers. The labeled substrate was then digested with R.NmeDI. After inactivating R.NmeDI, the reaction mixture was divided into two aliquots: one was mixed with stop solution immediately [lanes (−)]; the other was treated with Klenow Fragment at room temperature for 10 min and then mixed with stop solution (lane K). The R.NmeDI cleavage products were loaded onto 8% denatured polyacrylamide gel along with the products of the sequencing reaction based on the dideoxynucleotide chain termination method using pBluescript KS II (+) as the template and the same primers as for the extension reaction. (A) Determination of the cleavage site of R.NmeDI upstream of the recognition sequence. (B) Determination of the cleavage site of R.NmeDI downstream of the recognition sequence.