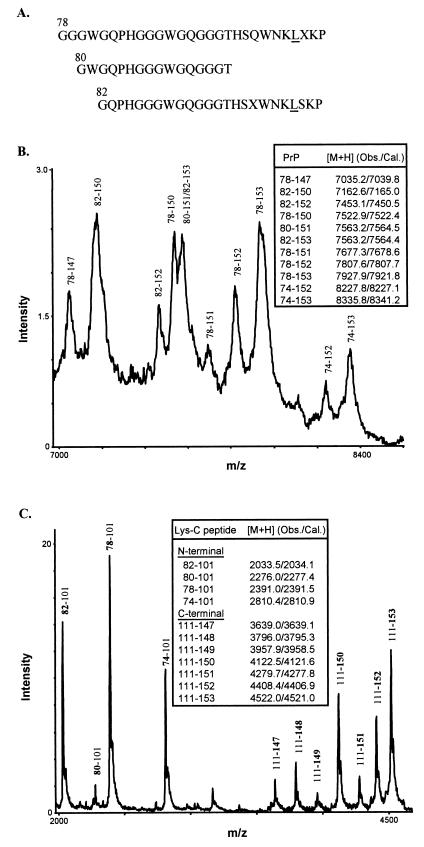
Figure 4.
Characterization of 8-kDa PrP-res by protein sequencing and mass spectrometry. (A) N-terminal sequences of purified 8-kDa PrP-res. The experimentally determined amino acid signals at each Edman degradation cycle were aligned with the known human PrP sequence to derive three major N-terminal species, with the residue positions shown above the first amino acids and the mutant residue at position 102 underlined. Unassigned residues were indicated as X. (B) Spectrum of 8-kDa PrP-res obtained by matrix-assisted laser desorption/ionization mass spectrometry. Major PrP species having Leu at position 102 and inclusive of N and C termini were assigned according to the observed and calculated protonated mass signals ([M+H] values shown in Inset). (C) Matrix-assisted laser desorption/ionization mass spectrum of the Lys-C digest of 8-kDa PrP-res. The N- and C-terminal PrP peptides generated by Lys-C digestion were assigned as in B. The N-terminal peptides corresponded to fragments that begin (bold) at positions 74, 78, 80, and 82, respectively, and end at the cleavage site at position 101 (the presence of Leu at position 102 introduces a specific cleavage at position 101). No wild-type N-terminal peptides were observed because, if present, they would correspond to PrP peptides having Pro at position 102 and spanning residues 74–106, 78–106, 80–106, and 82–106 (calculated [M+H] values 3348.6, 2929.2, 2815.1, 2571.8, respectively). The C-terminal peptides corresponded to fragments that begin at the cleavage site at positions 111 and end (bold) at positions 147, 148, 149, 150, 151, 152, and 153, respectively.