Fig. 1.
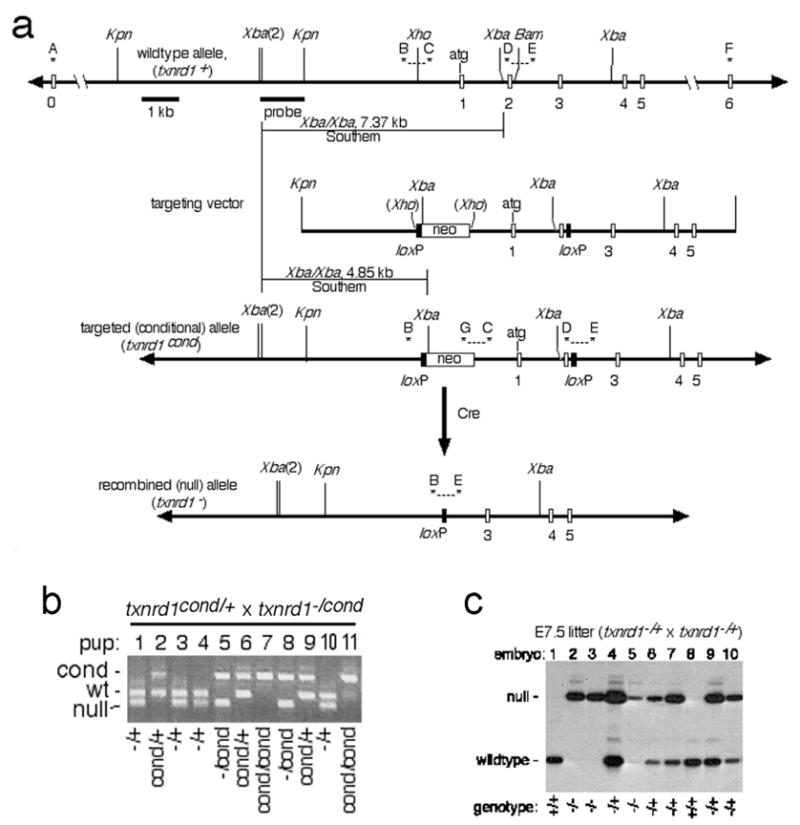
Design and confirmation of mice bearing txnrd1cond and txnrd1− alleles. a, schematic of alleles, targeting vector, and general mutagenesis strategy. Top, map of 5′ region of txnrd1 gene. Exons, represented as open boxes, are labeled below. Uppercase letters and asterisks indicate PCR primers. Probe and expected sizes of Southern blot bands are indicated. Below is the txnrd1-containing region of the targeting vector. Next is the correctly targeted (txnrd1cond) allele, with diagnostic PCR products and Southern blot bands indicated. Below this is the Cre-recombined null (txnrd1−) allele. b, confirmation of allele was determined by genetic analysis of mouse lines developed from ES cells. PCR-based genotyping reactions (primers B, C, E, G combined) were performed on genomic DNA isolated from tail snips at weaning from offspring of the indicated mating. This primer set gives the following allele-specific product sizes: txnrd1+, 338 bp; txnrd1cond, 542 bp; txnrd1−, 258 bp. Four pups had no txnrd1+ allele (pups 7 & 11 were txnrd1cond/cond; pups 5 & 8 were txnrd1−/cond), confirming that the downstream loxP was targeted into the txnrd1 allele. c, genotypic analysis of E7.5 embryos for microarray and RT-PCR analyses. Total nucleic acid was isolated from individual embryos and 10% of the total was used for radioactive PCR with primers B, D, and E. This primer set gives the following allele-specific product sizes: txnrd1+, 204 bp; txnrd1−, 258 bp.
