Fig. 6.
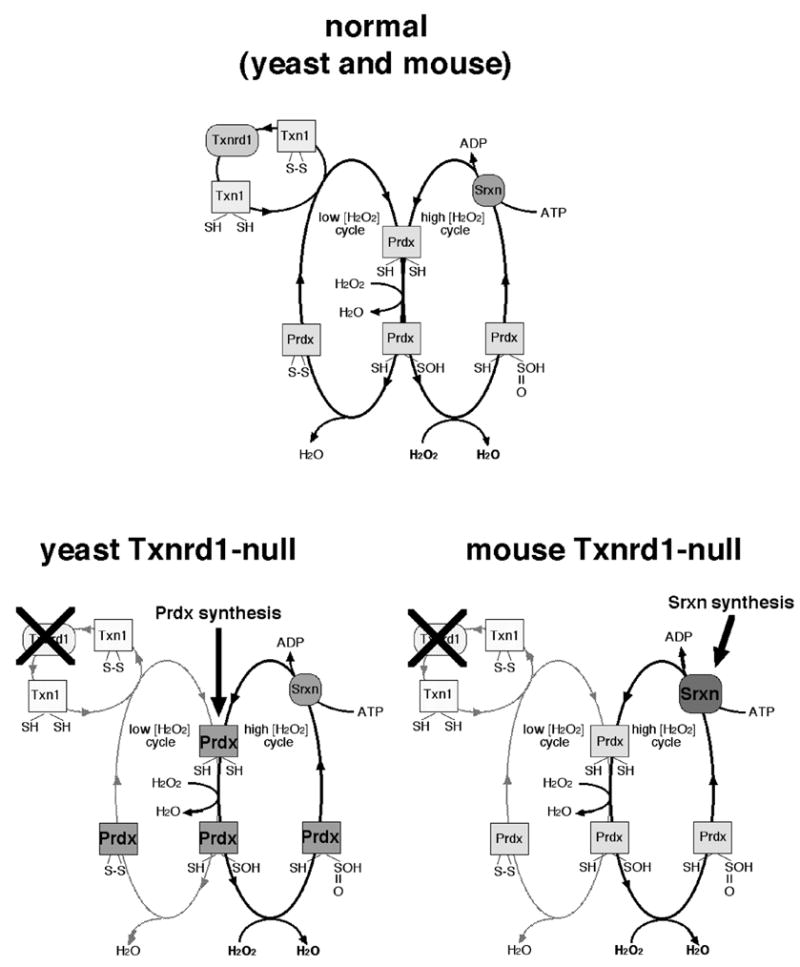
Eukaryotic Prdx catalytic cycles and inferred compensation for Txnrd1 disruption. At top is shown the normal Prdx catalytic cycles [68, 71]. The cycle at left is Txnrd1-dependent and functions to hydrolyze H2O2 at low concentrations. The cycle at right is Txnrd1-independent, and is only active at high H2O2 concentrations, where the Prdx sulfenic acid is further oxidized to a sulfinic acid, which cannot be reduced by Txn [74]. The mechanisms of Srxn catalysis are still under investigation; most models agree it is an ATP-dependent reaction [71, 72]. Below is shown the compensatory shifts expected in yeast [32] (left) and mice (right) in response to Txn system disruption. In both species, disruption of Txnrd1 is expected to disrupt the low [H2O2] Prdx cycle. Yeast strongly upregulate Prdx mRNA levels [32], suggesting compensation involves increasing rates of Prdx protein synthesis. By contrast, mice did not alter Prdx mRNA levels, but strongly upregulated Srxn mRNA (this study). Either response might bolster the high [H2O2] Prdx cycle.
