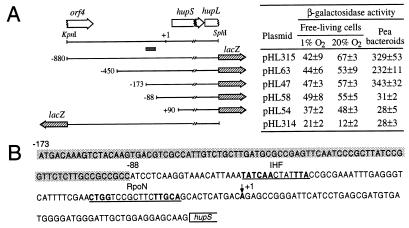
Figure 1.
Analysis of the hupS promoter. (A) Deletion analysis. The restriction and genetic maps of the promoter region are shown at the top left of the figure. The hupS transcriptional start site is indicated as position +1. 5′ deletion fragments were cloned in front of the promoterless lacZ gene in plasmid pMP220 (32) to generate plasmids of the pHL series. The position of the 5′ end with respect to the transcription start site is indicated. The hatched bar below the restriction map indicates the 85-bp DNA region required for hupSL transcription. β-galactosidase activities of aerobically and microaerobically grown cells and of pea bacteroids from R. leguminosarum bv. viciae UPM791 derivative strains carrying the pHL plasmids are shown on the right. Values (Miller units) are the average of three independent determinations ± SE. No differences in symbiotic stabilities of pHL plasmids were observed. (B) DNA sequence (GenBank/EMBL accession nos. U36924U36924 and X52974X52974). Regions similar to the RpoN (σ54) promoter consensus sequence YTGGCAC-N5-TTGCA (33) and to IHF-binding site WATCAA-N6-TTR (34) are underlined, and identical nucleotides are shown in boldface. The 85-bp region essential for hupSp activation is shadowed.