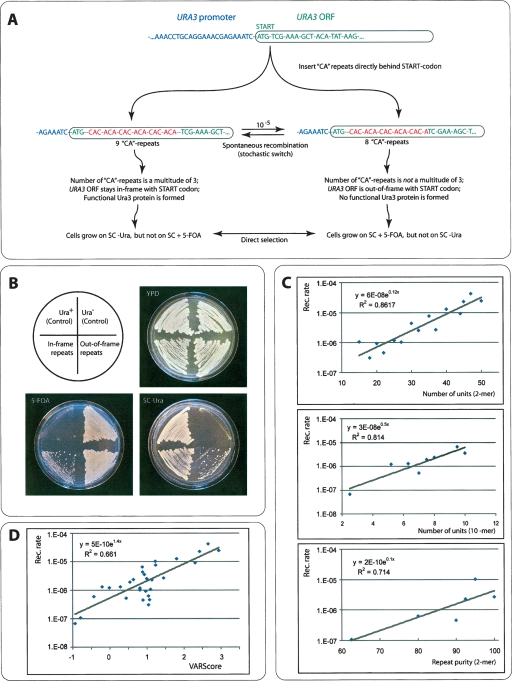
Figure 1.
VARscore correlates with repeat mutation rates. (A) To evaluate the correlation between the VARscore and experimentally determined mutation rates, a series of 30 different artificial repeats was inserted right behind the START codon of the genomic URA3 gene of a haploid S. cerevisiae S288C yeast strain. Three classes of strains were constructed: (1) a series of “CA” dinucleotide repeats with varying number of units; (2) a series of CA repeats with a constant number of units, but varying repeat purity; (3) a series of strains with a 10-mer and 20-mer unit length and varying number of units. (B) Since the number of nucleotides in each repeat is not a multitude of three, changes in the number of repeats lead to shifts in the URA3 reading frame, so that some strains will be Ura+, and others Ura−, depending on the number of repeat units they contain. Moreover, because of the instability of tandem repeats, the number of repeats will change in a fraction of each mitotic division, resulting in frequent shifts between Ura+ and URA− phenotypes. This can be demonstrated by growing cells in either SC–Ura or 5-FOA medium, which selects for Ura+ and Ura− strains, respectively (see Methods for details). (C) Plotting the mutation rates in the various repeat classes shows an exponential increase in mutation events with increasing unit number and purity. (D) Plotting VARscores for each repeat against their experimental mutation rates shows the correlation between VARscore and mutation rates, indicating that VARscores can be used as a rough estimation of mutation rate.