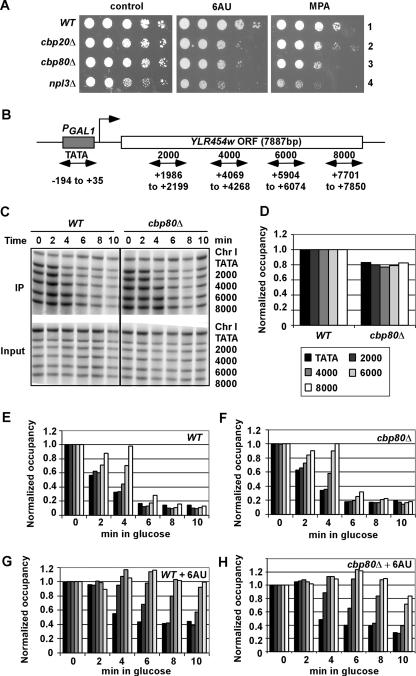
FIG. 4.
cbp80Δ does not detectably affect Pol II elongation rate and processivity in vivo. (A) cbp20Δ and npl3Δ mutants, like cbp80Δ cells, are sensitive to 6AU and MPA. Tenfold serial dilutions of strains BY4741 (WT), 2074 (cbp20Δ), 6566 (cbp80Δ), and 4268 (npl3Δ), transformed with YCplac33, were spotted on synthetic complete medium lacking uracil plates containing 2% glucose supplemented with 6AU (75 μg/ml) or MPA (15 μg/ml). (B) Diagram of the PGAL1-YLR454w gene with PCR primer pairs used to amplify the promoter and indicated coding sequences (numbered relative to ATG). (C) ChIP analysis of Rpb3p occupancies across the PGAL1-YLR454w gene induced with galactose and at different times of glucose repression. CMY017 (WT) or CMY018 (cbp80Δ) strains containing RPB3-myc and PGAL1-YLR454w were induced with galactose for 2 h (time = 0) and treated with 4% glucose for the indicated times. ChIP analysis was conducted using Rpb3p antibodies and primers (TATA to 8000, from top to bottom) shown in panel B. Amplification of a noncoding region on chromosome I (Chr I) was used as a control for nonspecific immunoprecipitation. (D) Quantification of the Rpb3p occupancies measured in panel C at the five indicated positions in WT and cbp80Δ cells grown in galactose medium (time = 0), normalized to the values in the WT strain at each position. (E and F) Quantification of the Rpb3p occupancies measured in panel C at the five positions indicated by the key in panel D in WT (E) or cbp80Δ (F) cells grown in glucose for the indicated times, normalized to the values in galactose (time = 0). (G and H) The assays were performed exactly as described for panels E and F except that cells were grown with 75 μg/ml 6AU. The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms.