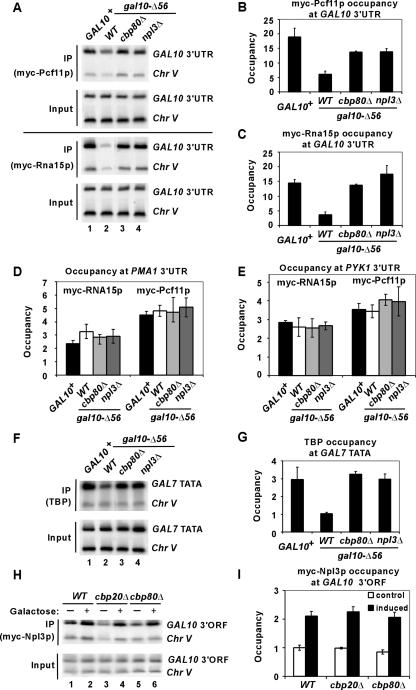
FIG. 6.
Cbp80p and Npl3p impede recruitment of polyadenylation/termination factors Pcf11p and Rna15p to the gal10-Δ56 terminator. (A to C) ChIP analysis of Myc-Pcf11p or Myc-Rna15p occupancies was conducted as described for Fig. 1E except using primers to amplify the GAL10 3′ UTR, employing PCF11-myc strains CMY019 (WT), CMY020 (gal10-Δ56), CMY021 (cbp80Δ gal10-Δ56), and CMY022 (npl3Δ gal10-Δ56) and RNA15-myc strains CMY023 (WT), CMY024 (gal10-Δ56), CMY025 (cbp80Δ gal10-Δ56), and CMY026 (npl3Δ gal10-Δ56). (D and E) Cbp80p and Npl3p do not impede recruitment of polyadenylation/termination factors Pcf11p and Rna15p to the PMA1 and PYK1 terminators. Myc-Pcf11p or Myc-Rna15p occupancies of the PMA1 and PYK1 3′ UTRs were analyzed in the same chromatin samples analyzed for panels A to C. (F and G) TATA binding protein (TBP) occupancies of the GAL7 TATA region were analyzed in the same chromatin samples analyzed for panels A to C. (H and I) Myc-Npl3p occupancy of the GAL10 3′ ORF is unaffected by cbp20Δ and cbp80Δ. ChIP analysis of Myc-Npl3p was done for NPL3-myc strains CMY027 (WT), CMY028 (cbp20Δ), and CMY029 (cbp80Δ) as described for Fig. 1E. Cells were cultured either in 2% raffinose (−) as a control or in 2% galactose (+) for induction of GAL10 expression. The average results obtained from two independent cultures and two PCR amplifications for each culture were plotted in the histograms with standard errors shown as error bars.