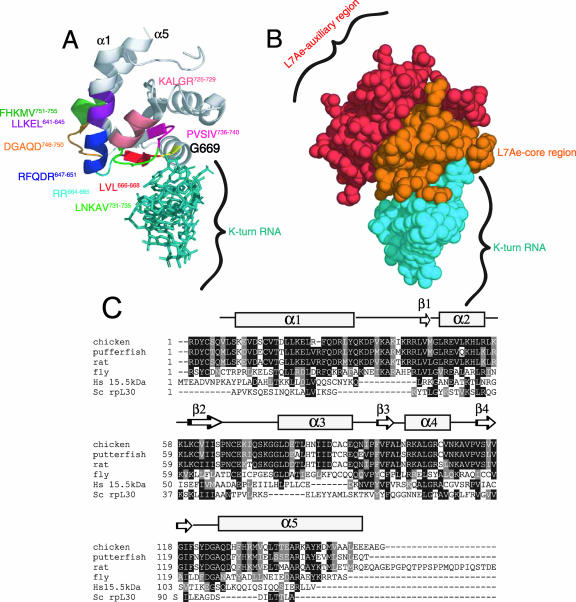
FIG. 6.
Structural modeling of SBP2. (A) The cocrystal structure of the 15.5-kDa snRNA binding protein bound to its cognate K-turn U4 RNA (Protein Data Bank coordinates 1E7K) was used to predict the positions of the following regions within CTSBP2: RFQDR647-651, DGAQD746-750, G669 (yellow), LVL666-668, RR664, KALGR725-729, LLKEL641-645, FHKMV752-756, LNKAV731-735, and PVSIV736-740. (B) Space-filled model of the structure presented in panel A, highlighting the auxiliary and core functional regions within the L7Ae domain as noted. The annotation of the structure was performed using MacPyMOL. (C) Alignment of the L7Ae motifs from the following K-turn binding proteins: Saccharomyces cerevisiae (Sc) rpL30, the human (Hs) 15.5-kDa protein, and the SBP2 sequences in Fig. 1. Residues shaded in black are identical, and residues shaded in gray are conservative substitutions. A secondary structure prediction for SBP2 (developed using PredictProtein) (26) is shown above the sequences.