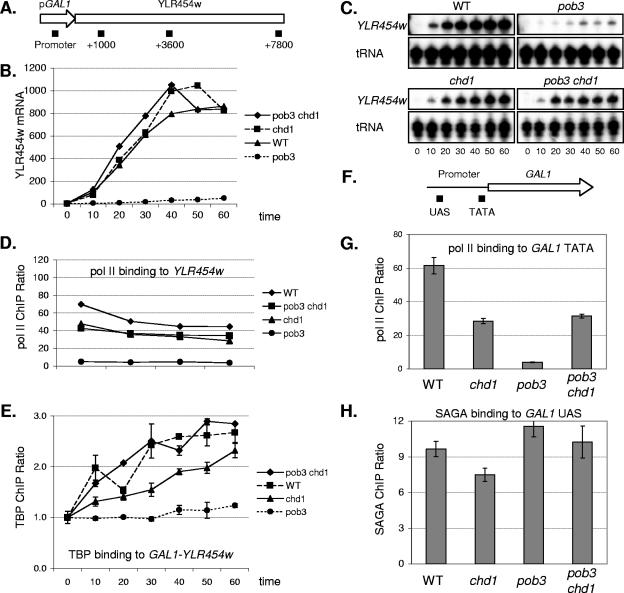
FIG. 7.
A chd1 mutation suppresses defects in GAL1 induction and Pol II and TBP binding caused by a pob3 mutation. Strains DY9591 (GAL1-YLR454w), DY9959 (chd1 GAL1-YLR454w), DY9972 (pob3 GAL1-YLR454w), and DY10020 (pob3 chd1 GAL1-YLR454w) were grown on YP medium with 2% raffinose. Galactose was added to 2%, and samples were taken at 10-min intervals and processed for ChIP analysis to measure Pol II and TBP binding. (A) Map of the GAL1-YLR454w allele showing the positions of regions amplified by at the promoter and within the gene. (B) YLR454w mRNA levels measured from the GAL1-YLR454w allele, quantified after phosphorimaging of the gels in panel C. WT, wild type. (C) S1 protection assays to measure YLR454w mRNA from the GAL1-YLR454w allele, using probes specific for YLR454w and a tRNA internal control. (D) Distribution of Pol II at 60 min following galactose induction at different GAL1-YLR454w regions in four different strains. Error bars show variance (standard deviations) among replicate PCRs. (E) TBP binding to the GAL1-YLR454w promoter following galactose induction in four different strains. ChIP values were normalized to binding at time zero. Error bars show variance among replicate PCRs. (F) Map of the native GAL1 gene showing the positions of regions amplified by at the upstream activation sequence (UAS) and TATA within the promoter. (G) Pol II binding to the TATA region (positions −190 to + 54) of the native GAL1 promoter at 30 min following galactose induction in four different strains. Error bars show variance among replicate PCRs. (H) SAGA binding to the native UAS region (positions −496 to −316) of the native GAL1 promoter at 30 min following galactose induction in four different strains. Error bars show variance among replicate PCRs.