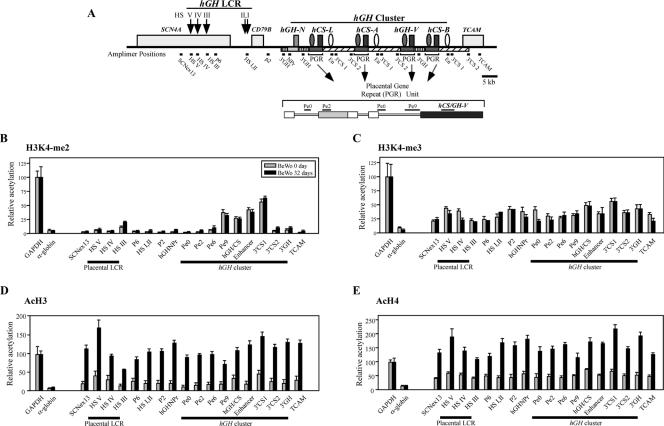
FIG. 5.
Histone modifications at the hGH cluster and its LCR in BeWo cells during hGH-hCS activation. (A) Positions of amplimers used in the ChIP assays. The structure of the hGH cluster, linked genes, and the hGH LCR are depicted. P-elements and the putative hCS enhancers are indicated (shaded and open ovals, respectively). Shaded and striped rectangles indicate extensive conserved segments among the PGR units and the hGH-N gene. The amplimer names and their positions are shown below the map. The PGR unit is also shown in an expanded format. (B) The histone H3K4 dimethylation pattern in BeWo cells was established prior to gene activation and did not change after long-term culture. Soluble chromatin was collected from BeWo cells at days 0 and 32 of culture by digesting nuclei with micrococcal nuclease. The ChIP analysis was conducted with anti-dimethylated-histone H3K4 antibody, and the resulting input and bound fractions were subjected to PCR amplification. The bound/input signal ratio at each amplicon (A) was normalized to that at the GAPDH promoter, which was considered 100. The relative histone modification levels in day 0 (shaded bars) and day 32 (black rectangles) BeWo cells are shown. Standard error values are indicated. The α-globin promoter was used as a negative control. (C) Histone H3K4 trimethylation was detected at low levels across the entire locus in BeWo cells at both day 0 and day 32. Histone H3K4 trimethylation patterns, examined by ChIP analysis with anti-trimethylated-H3K4 antibody, were analyzed and plotted as described for panel B. (D) Histone H3 acetylation was significantly induced across the locus during the 32 days of BeWo cell culture. Histone H3 acetylation patterns, using anti-acetylated-histone H3 antibody, were studied as described for panel B. (E) Histone H4 acetylation was significantly induced across the locus during BeWo cell culture. The ChIP assay was performed, analyzed, and plotted as described for panel B but with anti-acetyl histone H4 antibody.