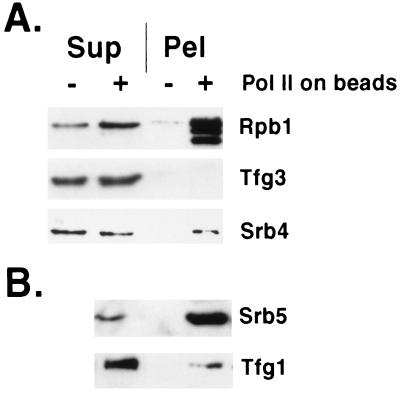
Figure 1.
Mediator exchange between RNA polymerases. Exchange was for 10 min (A) or 20 min (B, separate experiment from A) by slow rotation at room temperature between free RNA polymerase II holoenzyme (0.1–0.5 μg) and hexahistidine-tagged core RNA polymerase II bound to Ni2+-agarose beads (+) or beads with no bound polymerase (−). Following sedimentation in a microcentrifuge for 2 min at 5,000 rpm, supernatant (Sup) and beads (Pel) were analyzed by SDS/PAGE (10% gel) and immunoblotting with antibodies against the proteins indicated. The Rpb1 subunit of the polymerase bound to beads was partly N-terminally degraded, resulting in a double band, which served to identify the core polymerase. Binding of core polymerase (25–50 μg in 40–150 μl) to beads (Qiagen) was in 40 mM Hepes, pH 7.6/20% glycerol/5 mM 2-mercaptoethanol/0.25 mg/ml bovine serum albumin/protease inhibitors (12) by slow rotation for 45 min at 4°C. Following several washes by centrifugation and resuspension in 200 mM Hepes, pH 7.6/20% glycerol/5 mM 2-mercaptoethanol/0.25 mg/ml bovine serum albumin/0.2% Tween-20/0.01% Nonidet P-40/10 mM imidazole/protease inhibitors, beads were resuspended in 100 μl of the same buffer. Exchange was performed with 10 μl of beads (2–5 μg of bound polymerase) in 40 mM Hepes, pH 7.6/150 mM potassium acetate/5 mM magnesium acetate/10% glycerol/5 mM 2-mercaptoethanol/0.1 mg/ml bovine serum albumin/0.06% Tween-20/0.003% Nonidet P-40/3 mM imidazole/protease inhibitors.