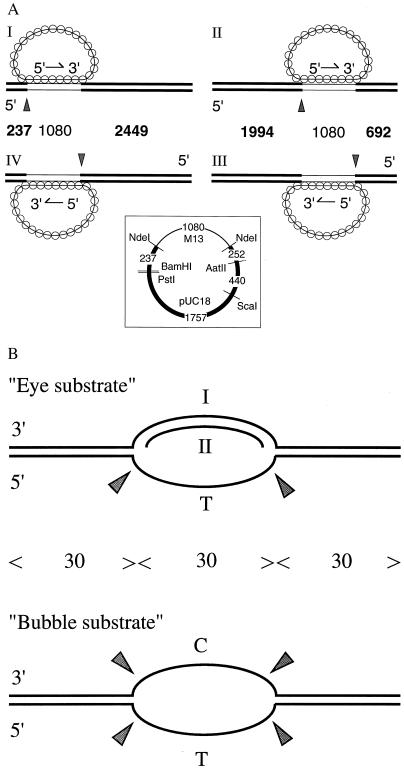
Figure 1.
Substrates of the junction endonuclease. (A) Paranemic joints were made by pairing RecA filaments containing circular single-stranded M13 DNA with an ≈1-kb homologous region in linear duplex DNA that was labeled at its 5′ ends. RecA nucleoprotein filaments were formed on circular single-stranded M13 DNA, called the viral or plus strand. The strand in the duplex containing the same sequence as the single strand in the filament is called the anticomplementary strand. Linear duplex DNA for substrates I and IV was made by cleaving the vector derived from pUC18 with BamHI, and for substrates II and III, by cleaving with ScaI. Substrate I was the same as substrate IV except that the ≈1-kb homologous region was reversed in orientation; the same was true for substrates II vs. III. Thin lines in the diagrams of duplex DNA indicate regions that are homologous to M13. The numbers between the diagrams mark the lengths in base pairs of the indicated regions; the numbers in boldface indicate fragments produced by junction endonuclease acting at the points marked by arrowheads. (Inset) A map of the pUC18 vector carrying the ≈1-kb segment of M13. The numbers indicate base pairs of the designated regions. (B) Synthetic oligonucleotide substrates that were cleaved by junction endonuclease in the absence of RecA protein. See Materials and Methods for details of preparation. C and T mark homopolymeric regions; all other regions contained all four bases. The numbers of nucleotide residues are indicated between the diagrams. Sites of cleavage are marked by arrowheads.