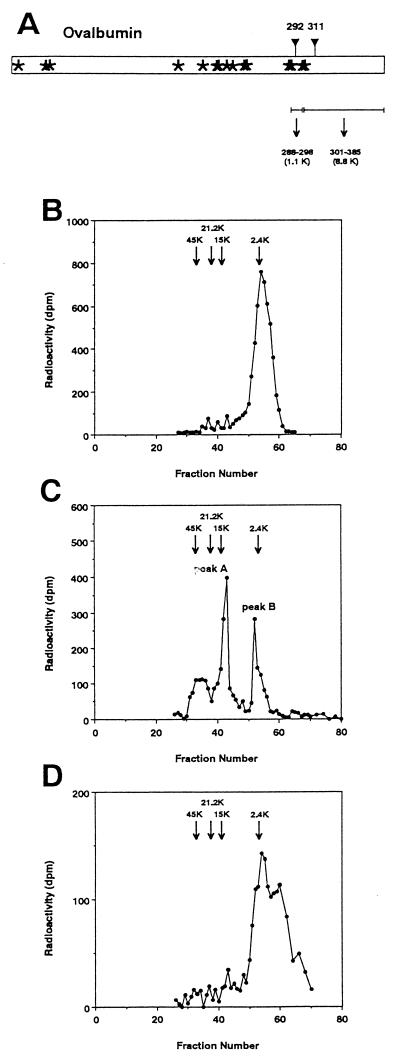
Figure 2.
Determination of the site of de-N-glycosylation after treatment of Di-OVA with PNGase HO. (a) Schematic drawing showing cleavage of OVA by CNBr treatment. Asterisks show methionine residues. ▿, Site of potential N-glycosylation sites, Asn-292 and Asn-311. The molecular sizes of the peptide moieties of the CNBr fragments involving the potential N-glycosylation sites are as follows: peptide 288–298, 1.1 kDa; and peptide 301–385, 8.8 kDa. Sephadex G-50 chromatography of CNBr fragments from OE (b), Di-OVA (c), and Mono-OVA obtained by PNGase HO-treatment of Di-OVA (d). Arrows in the chromatograms denote positions of the marker proteins: OVA (45 kDa), soybean trypsin inhibitor (21 kDa), ribonuclease B (15 kDa), and high-mannose-type glycopeptide from OVA (2.4 kDa).