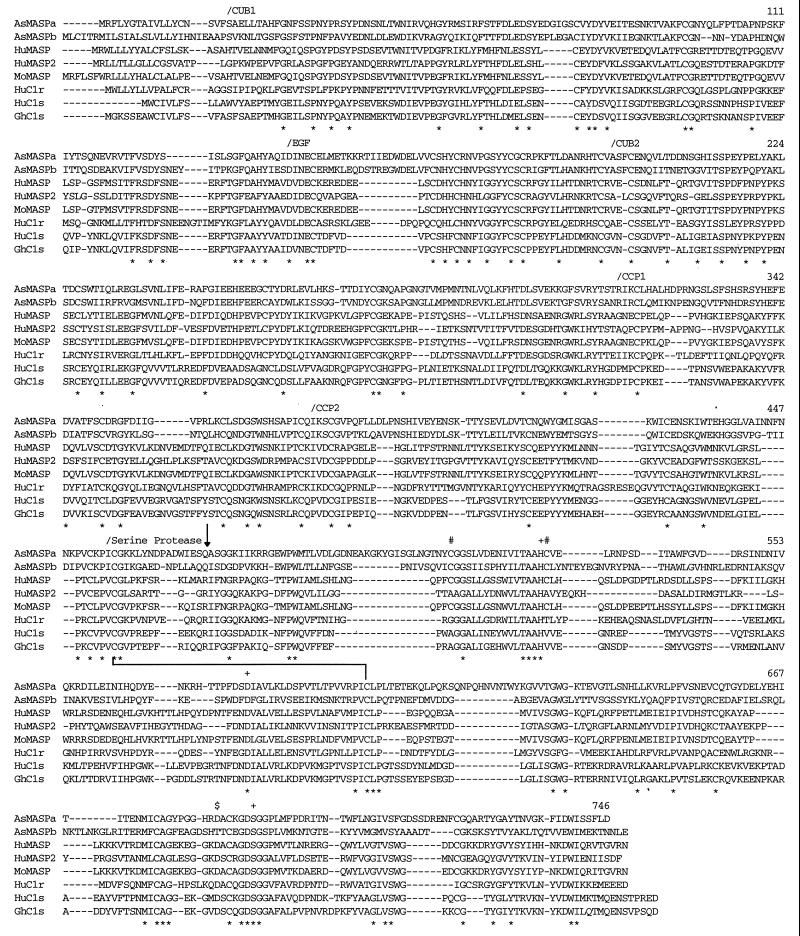
Figure 3.
Alignment of AsMASPa and AsMASPb amino acid sequences with mammalian MASP, C1r, and C1s. Multiple alignment of the amino acid sequences was performed using Clustal W software. Gaps introduced to increase identity are shown by dashes. The completely conserved residues among these eight sequences are indicated by ∗ below the sequences. The amino acid numbers of the rightmost residues of AsMASPa are shown for each lane. The N-termini of each domain are marked by/above the sequences. Three residues at the catalytic site of serine proteases 527H, 579D, and 690S, two cysteine residues involved in histidine loop formation (512C and 528C), and the residue at the S1 specificity crevice (684D) are marked by +, #, and $, respectively. The arrow pointing downward indicates a processing site of mammalian MASP, C1r, and C1s. The line connecting 456C and 599C indicates a disulfide bond between two subunit chains.