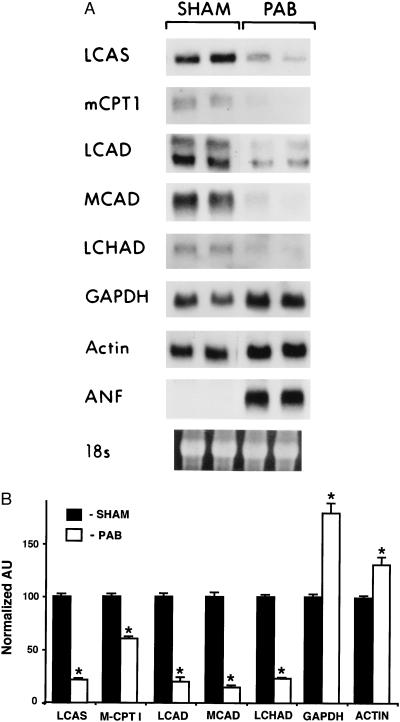
Figure 1.
Expression of genes involved in myocardial fatty acid utilization and glycolysis parallels known changes in substrate preferences in the hypertrophied heart. (A) Representative Northern blot analysis obtained with total RNA isolated from the RV of two mice subjected to PAB or from sham-operated controls (SHAM). Each lane contains 15 μg of total RNA. cDNA probe abbreviations are defined in the text. The 18S rRNA (stained with ethidium bromide) within each lane is shown at the bottom. (B) Densitometric analysis of Northern blots obtained with RNA isolated from the RV of six PAB or six sham-operated control mice. The bars represent mean (± SEM) steady-state mRNA levels (in arbitrary units or AU) normalized to sham-operated control values (mean control value set at 100). ∗ denotes P < 0.01 compared with the corresponding control value. LCAD, long-chain acyl-CoA dehydrogenase; LCAS, long-chain acyl-CoA synthetase; MCPT1, muscle isoform of carnitine palmitoyltransferase I; LCHAD, long-chain 3-OH acyl-CoA dehydrogenase.