Abstract
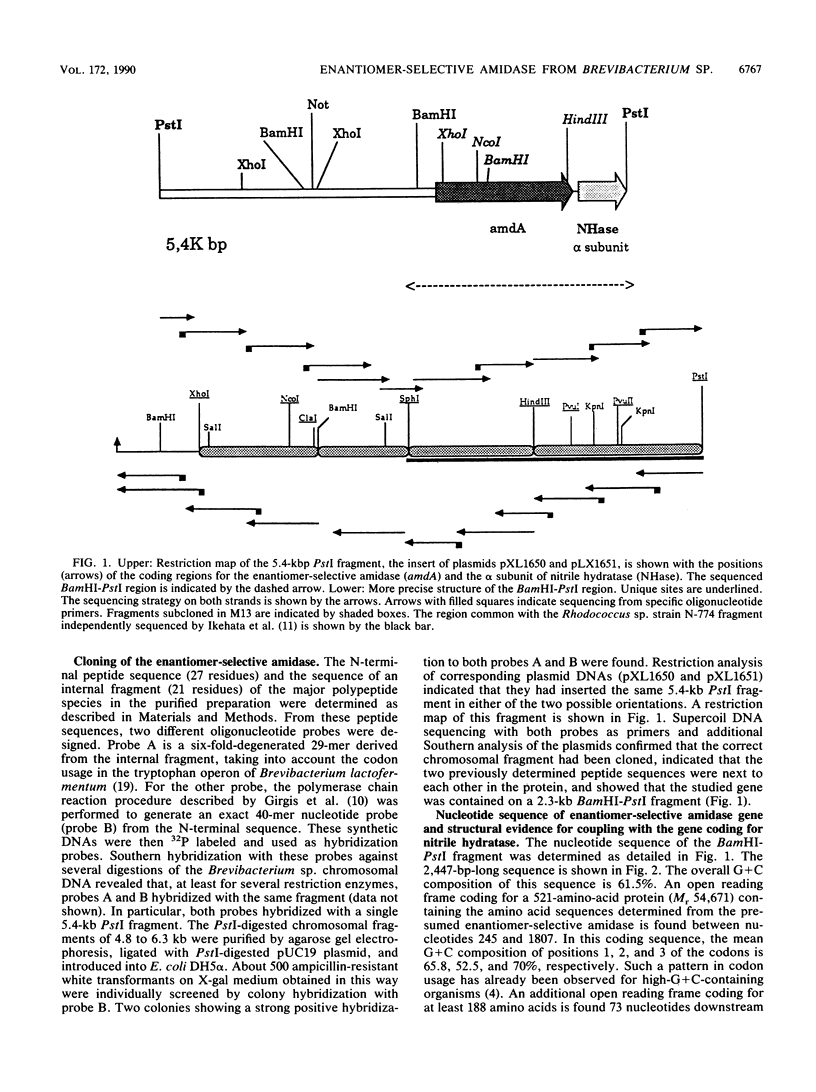
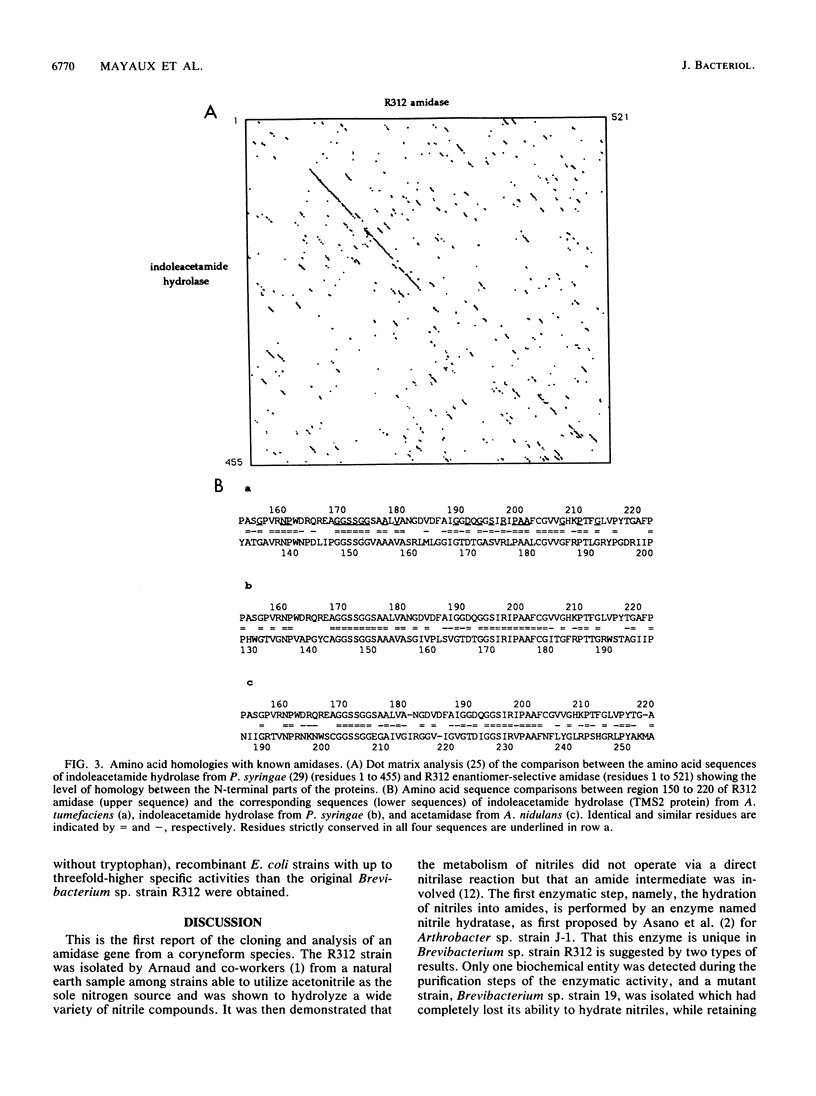
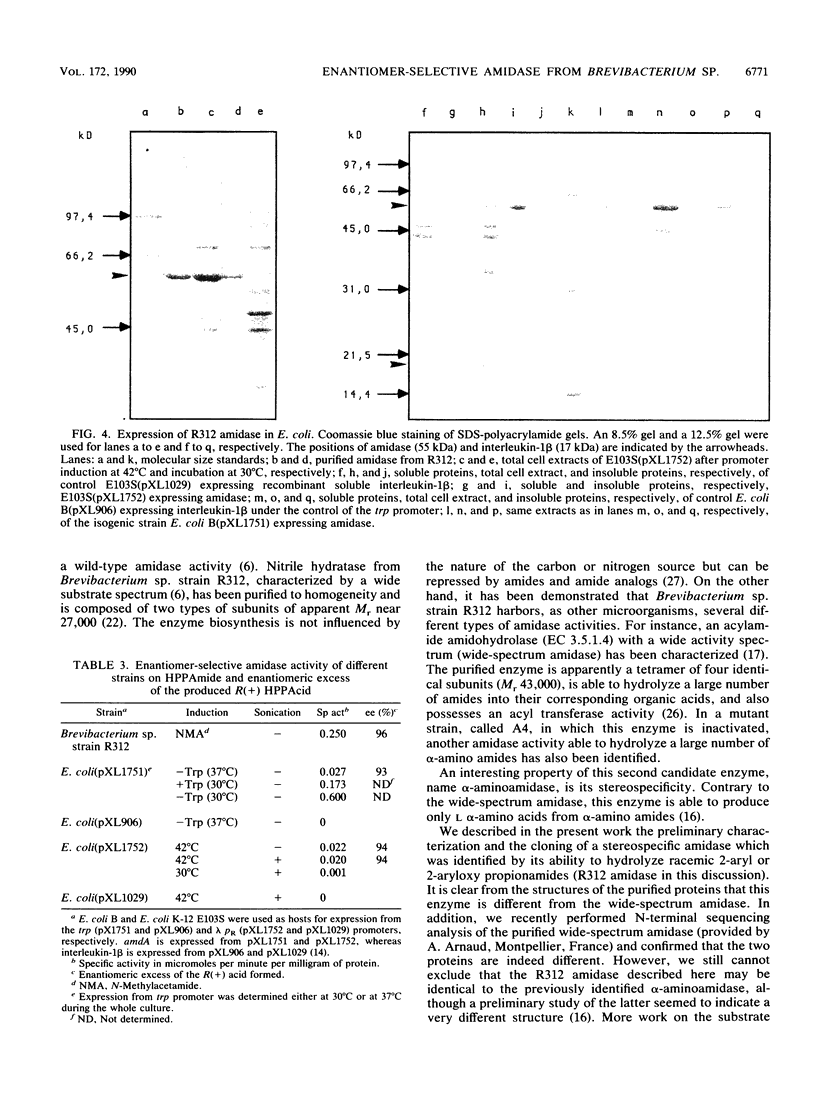
An enantiomer-selective amidase active on several 2-aryl and 2-aryloxy propionamides was identified and purified from Brevibacterium sp. strain R312. Oligonucleotide probes were designed from limited peptide sequence information and were used to clone the corresponding gene, named amdA. Highly significant homologies were found at the amino acid level between the deduced sequence of the enantiomer-selective amidase and the sequences of known amidases such as indoleacetamide hydrolases from Pseudomonas syringae and Agrobacterium tumefaciens and acetamidase from Aspergillus nidulans. Moreover, amdA is found in the same orientation and only 73 bp upstream from the gene coding for nitrile hydratase, strongly suggesting that both genes are part of the same operon. Our results also showed that Rhodococcus sp. strain N-774 and Brevibacterium sp. strain R312 are probably identical, or at least very similar, microorganisms. The characterized amidase is an apparent homodimer of Mr 2 x 54,671 which exhibited under our conditions a specific activity of about 13 to 17 mumol of 2-(4-hydroxyphenoxy)propionic R acid formed per min per mg of enzyme from the racemic amide. Large amounts of an active recombinant enzyme could be produced in Escherichia coli at 30 degrees C under the control of an E. coli promoter and ribosome-binding site.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arnaud A., Galzy P., Jallageas J. C. Remarques sur l'activité nitrilasique de quelques bactéries. C R Acad Sci Hebd Seances Acad Sci D. 1976 Sep 20;283(5):571–573. [PubMed] [Google Scholar]
- Bibb M. J., Findlay P. R., Johnson M. W. The relationship between base composition and codon usage in bacterial genes and its use for the simple and reliable identification of protein-coding sequences. Gene. 1984 Oct;30(1-3):157–166. doi: 10.1016/0378-1119(84)90116-1. [DOI] [PubMed] [Google Scholar]
- Corrick C. M., Twomey A. P., Hynes M. J. The nucleotide sequence of the amdS gene of Aspergillus nidulans and the molecular characterization of 5' mutations. Gene. 1987;53(1):63–71. doi: 10.1016/0378-1119(87)90093-x. [DOI] [PubMed] [Google Scholar]
- Denèfle P., Kovarik S., Guitton J. D., Cartwright T., Mayaux J. F. Chemical synthesis of a gene coding for human angiogenin, its expression in Escherichia coli and conversion of the product into its active form. Gene. 1987;56(1):61–70. doi: 10.1016/0378-1119(87)90158-2. [DOI] [PubMed] [Google Scholar]
- Follettie M. T., Sinskey A. J. Molecular cloning and nucleotide sequence of the Corynebacterium glutamicum pheA gene. J Bacteriol. 1986 Aug;167(2):695–702. doi: 10.1128/jb.167.2.695-702.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis S. I., Alevizaki M., Denny P., Ferrier G. J., Legon S. Generation of DNA probes for peptides with highly degenerate codons using mixed primer PCR. Nucleic Acids Res. 1988 Nov 11;16(21):10371–10371. doi: 10.1093/nar/16.21.10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehata O., Nishiyama M., Horinouchi S., Beppu T. Primary structure of nitrile hydratase deduced from the nucleotide sequence of a Rhodococcus species and its expression in Escherichia coli. Eur J Biochem. 1989 May 15;181(3):563–570. doi: 10.1111/j.1432-1033.1989.tb14761.x. [DOI] [PubMed] [Google Scholar]
- Jallageas J. C., Arnaud A., Galzy P. Remarques sur le spectre d'activité amidasique d'un mutant de Brevibacterium. C R Seances Acad Sci D. 1979 Feb 12;288(6):655–658. [PubMed] [Google Scholar]
- Jung G., Denèfle P., Becquart J., Mayaux J. F. High-cell density fermentation studies of recombinant Escherichia coli strains expressing human interleukin-1 beta. Ann Inst Pasteur Microbiol. 1988 Jan-Feb;139(1):129–146. doi: 10.1016/0769-2609(88)90100-7. [DOI] [PubMed] [Google Scholar]
- Kanehisa M. Use of statistical criteria for screening potential homologies in nucleic acid sequences. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):203–213. doi: 10.1093/nar/12.1part1.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latta M., Philit M., Maury I., Soubrier F., Denèfle P., Mayaux J. F. Tryptophan promoter derivatives on multicopy plasmids: a comparative analysis of expression potentials in Escherichia coli. DNA Cell Biol. 1990 Mar;9(2):129–137. doi: 10.1089/dna.1990.9.129. [DOI] [PubMed] [Google Scholar]
- Matsui K., Sano K., Ohtsubo E. Sequence analysis of the Brevibacterium lactofermentum trp operon. Mol Gen Genet. 1987 Sep;209(2):299–305. doi: 10.1007/BF00329657. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Nanba H., Ryuno K., Takeuchi K., Yamada H. Nitrile hydratase of Pseudomonas chlororaphis B23. Purification and characterization. Eur J Biochem. 1987 Feb 2;162(3):691–698. doi: 10.1111/j.1432-1033.1987.tb10692.x. [DOI] [PubMed] [Google Scholar]
- Nagasawa T., Ryuno K., Yamada H. Nitrile hydratase of Brevibacterium R312--purification and characterization. Biochem Biophys Res Commun. 1986 Sep 30;139(3):1305–1312. doi: 10.1016/s0006-291x(86)80320-5. [DOI] [PubMed] [Google Scholar]
- Schröder G., Waffenschmidt S., Weiler E. W., Schröder J. The T-region of Ti plasmids codes for an enzyme synthesizing indole-3-acetic acid. Eur J Biochem. 1984 Jan 16;138(2):387–391. doi: 10.1111/j.1432-1033.1984.tb07927.x. [DOI] [PubMed] [Google Scholar]
- Staden R. Methods to define and locate patterns of motifs in sequences. Comput Appl Biosci. 1988 Mar;4(1):53–60. doi: 10.1093/bioinformatics/4.1.53. [DOI] [PubMed] [Google Scholar]
- Tourneix D., Thiéry A., Maestracci M., Arnaud A., Galzy P. Regulation of nitrile-hydratase synthesis in a Brevibacterium species. Antonie Van Leeuwenhoek. 1986;52(2):173–182. doi: 10.1007/BF00429321. [DOI] [PubMed] [Google Scholar]
- Yamada T., Palm C. J., Brooks B., Kosuge T. Nucleotide sequences of the Pseudomonas savastanoi indoleacetic acid genes show homology with Agrobacterium tumefaciens T-DNA. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6522–6526. doi: 10.1073/pnas.82.19.6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]



