Abstract
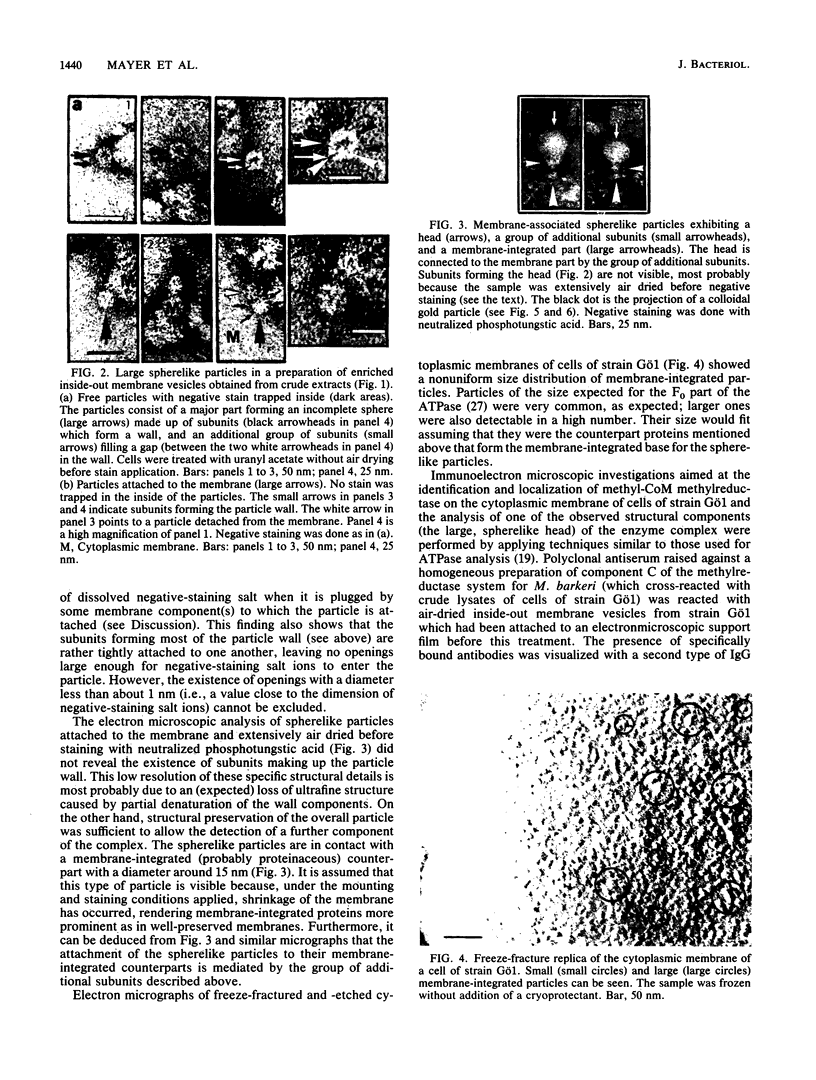
The methanogenic bacterium strain Gö1 harbors a high-molecular-weight enzyme complex containing methyl coenzyme M methylreductase as revealed by immunoelectron microscopy. This complex consists of a spherelike, hollow head piece, in the wall of which a number of copies of the methyl coenzyme M methylreductase are located. It is named Rc (c indicates collector). Intimately bound to it is a group of additional subunits of unknown composition referred to as Rm (m indicates mediator). Electron microscopy of negatively stained samples indicated that Rm contains a functional pore or channel which connects the internal volume of Rc with the outside. The RcRm complex is named Rs (s indicates spherelike). This complex was often found detached from the inside of the cytoplasmic membrane when membrane vesicles were investigated. However, Rs was also seen attached to a third component of the complex located in the membrane, the attachment being mediated by Rm. This membrane part of the complex is designated Rt (t indicates translocator). It consists of subunits with unknown composition. When Rs is attached to the membrane, the pore in Rm appears to be plugged by Rt. This indicates that the internal volume in Rc is in contact, via the pore in Rm, with Rt. The RcRmRt complex is referred to as methanoreductosome. Functional implications of the structural organization of the methylreductase system are discussed in view of methane formation and the creation of a transmembrane proton gradient used by the cell for ATP synthesis.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Balch W. E., Fox G. E., Magrum L. J., Woese C. R., Wolfe R. S. Methanogens: reevaluation of a unique biological group. Microbiol Rev. 1979 Jun;43(2):260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaut M., Gottschalk G. Coupling of ATP synthesis and methane formation from methanol and molecular hydrogen in Methanosarcina barkeri. Eur J Biochem. 1984 May 15;141(1):217–222. doi: 10.1111/j.1432-1033.1984.tb08178.x. [DOI] [PubMed] [Google Scholar]
- De Mey J., Moeremans M., Geuens G., Nuydens R., De Brabander M. High resolution light and electron microscopic localization of tubulin with the IGS (immuno gold staining) method. Cell Biol Int Rep. 1981 Sep;5(9):889–899. doi: 10.1016/0309-1651(81)90204-6. [DOI] [PubMed] [Google Scholar]
- Ellefson W. L., Wolfe R. S. Component C of the methylreductase system of Methanobacterium. J Biol Chem. 1981 May 10;256(9):4259–4262. [PubMed] [Google Scholar]
- Gunsalus R. P., Wolfe R. S. Methyl coenzyme M reductase from Methanobacterium thermoautotrophicum. Resolution and properties of the components. J Biol Chem. 1980 Mar 10;255(5):1891–1895. [PubMed] [Google Scholar]
- Hippe H., Caspari D., Fiebig K., Gottschalk G. Utilization of trimethylamine and other N-methyl compounds for growth and methane formation by Methanosarcina barkeri. Proc Natl Acad Sci U S A. 1979 Jan;76(1):494–498. doi: 10.1073/pnas.76.1.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannssen W., Schütte H., Mayer F., Mayer H. Quaternary structure of the isolated restriction endonuclease EndoR.Bgl I from Bacillus globigii as revealed by electron microscopy. J Mol Biol. 1979 Nov 15;134(4):707–726. doi: 10.1016/0022-2836(79)90481-9. [DOI] [PubMed] [Google Scholar]
- Knecht E., Martinez-Ramon A., Grisolia S. Electron microscopic localization of glutamate dehydrogenase in rat liver mitochondria by an immunogold procedure and monoclonal and polyclonal antibodies. J Histochem Cytochem. 1986 Jul;34(7):913–922. doi: 10.1177/34.7.3519755. [DOI] [PubMed] [Google Scholar]
- Kohring G. W., Mayer F., Mayer H. Immunoelectron microscopic localization of the restriction endonuclease EcoRI in Escherichia coli BS 5. Eur J Cell Biol. 1985 May;37:1–6. [PubMed] [Google Scholar]
- Mayer F., Jussofie A., Salzmann M., Lübben M., Rohde M., Gottschalk G. Immunoelectron microscopic demonstration of ATPase on the cytoplasmic membrane of the methanogenic bacterium strain Göl. J Bacteriol. 1987 May;169(5):2307–2309. doi: 10.1128/jb.169.5.2307-2309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer F., Wallace J. C., Keech D. B. Further electron microscope studies on pyruvate carboxylase. Eur J Biochem. 1980 Nov;112(2):265–272. doi: 10.1111/j.1432-1033.1980.tb07202.x. [DOI] [PubMed] [Google Scholar]
- Nagle D. P., Jr, Wolfe R. S. Component A of the methyl coenzyme M methylreductase system of Methanobacterium: resolution into four components. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2151–2155. doi: 10.1073/pnas.80.8.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossmer R., Mund T., Hartzell P. L., Konheiser U., Kohring G. W., Klein A., Wolfe R. S., Gottschalk G., Mayer F. Immunocytochemical localization of component C of the methylreductase system in Methanococcus voltae and Methanobacterium thermoautotrophicum. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5789–5792. doi: 10.1073/pnas.83.16.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirola B. A., Mayer F., Borthwick I. A., Srivastava G., May B. K., Elliott W. H. Electron microscopic studies on liver 5-aminolaevulinate synthase. Eur J Biochem. 1984 Nov 2;144(3):577–579. doi: 10.1111/j.1432-1033.1984.tb08504.x. [DOI] [PubMed] [Google Scholar]
- Shapiro S., Wolfe R. S. Methyl-coenzyme M, an intermediate in methanogenic dissimilation of C1 compounds by Methanosarcina barkeri. J Bacteriol. 1980 Feb;141(2):728–734. doi: 10.1128/jb.141.2.728-734.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot J. W., Geuze H. J. Sizing of protein A-colloidal gold probes for immunoelectron microscopy. J Cell Biol. 1981 Aug;90(2):533–536. doi: 10.1083/jcb.90.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor C. D., Wolfe R. S. Structure and methylation of coenzyme M(HSCH2CH2SO3). J Biol Chem. 1974 Aug 10;249(15):4879–4885. [PubMed] [Google Scholar]
- Tiedge H., Schäfer G., Mayer F. An electron microscopic approach to the quaternary structure of mitochondrial F1-ATPase. Eur J Biochem. 1983 Apr 15;132(1):37–45. doi: 10.1111/j.1432-1033.1983.tb07322.x. [DOI] [PubMed] [Google Scholar]
- Valentine R. C., Shapiro B. M., Stadtman E. R. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry. 1968 Jun;7(6):2143–2152. doi: 10.1021/bi00846a017. [DOI] [PubMed] [Google Scholar]
- Wackett L. P., Hartwieg E. A., King J. A., Orme-Johnson W. H., Walsh C. T. Electron microscopy of nickel-containing methanogenic enzymes: methyl reductase and F420-reducing hydrogenase. J Bacteriol. 1987 Feb;169(2):718–727. doi: 10.1128/jb.169.2.718-727.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]









