Abstract
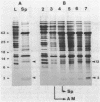
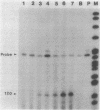
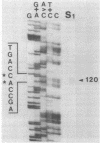
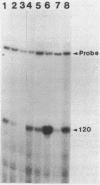
Streptomyces coelicolor is a filamentous, gram-positive bacterium that exhibits a complex cycle of morphological differentiation involving the formation of an aerial mycelium of multinucleoid hyphae which undergo septation to form long chains of spores. We report the identification of two proteins of 13 and 3 kilodaltons, designated SapA and SapB, respectively, that are produced during formation of the aerial mycelium and are found in assocation with purified, mature spores. We cloned the structural gene (sapA) for one of these spore-associated proteins. Nucleotide sequence analysis suggests that the 13-kilodalton polypeptide is derived from a larger pre- or preproprotein containing a leader sequence of 37 amino acids. Nuclease protection-hybridization analysis and experiments using the Vibrio harveyi, luciferase-encoding luxAB operon as a gene tag demonstrated that expression of sapA is controlled from a promoter contained within a region of less than 110 base pairs in length, whose transcription start site is located approximately 50 base pairs upstream from the initiation codon for the sapA open reading frame. Transcription of sapA was induced at the time of appearance of the aerial mycelium, and the level of sapA transcripts was significantly reduced in certain mutants blocked in aerial mycelium (bld) and or spore (whi) formation. As further evidence of the association of sapA transcription with morphological differentiation, experiments in which we monitored sapA transcription topographically by use of a sapA-luxAB operon fusion demonstrated a close spatial correlation between colony regions undergoing aerial mycelium formation and zones of sapA-promoted light emission.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Binnie C., Lampe M., Losick R. Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater K. F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1972 Aug;72(1):9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- Deng Z., Kieser T., Hopwood D. A. Expression of a Streptomyces plasmid promoter in Escherichia coli. Gene. 1986;43(3):295–300. doi: 10.1016/0378-1119(86)90219-2. [DOI] [PubMed] [Google Scholar]
- Donovan W., Zheng L. B., Sandman K., Losick R. Genes encoding spore coat polypeptides from Bacillus subtilis. J Mol Biol. 1987 Jul 5;196(1):1–10. doi: 10.1016/0022-2836(87)90506-7. [DOI] [PubMed] [Google Scholar]
- Goldman R. C., Tipper D. J. Bacillus subtilis spore coats: complexity and purification of a unique polypeptide component. J Bacteriol. 1978 Sep;135(3):1091–1106. doi: 10.1128/jb.135.3.1091-1106.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A. Genetic analysis and genome structure in Streptomyces coelicolor. Bacteriol Rev. 1967 Dec;31(4):373–403. doi: 10.1128/br.31.4.373-403.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunkapiller M. W., Lujan E., Ostrander F., Hood L. E. Isolation of microgram quantities of proteins from polyacrylamide gels for amino acid sequence analysis. Methods Enzymol. 1983;91:227–236. doi: 10.1016/s0076-6879(83)91019-4. [DOI] [PubMed] [Google Scholar]
- Igo M. M., Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986 Oct 20;191(4):615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mendez C., Chater K. F. Cloning of whiG, a gene critical for sporulation of Streptomyces coelicolor A3(2). J Bacteriol. 1987 Dec;169(12):5715–5720. doi: 10.1128/jb.169.12.5715-5720.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick M. J. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1976 Oct;96(2):299–315. doi: 10.1099/00221287-96-2-299. [DOI] [PubMed] [Google Scholar]
- Mills D. R., Kramer F. R. Structure-independent nucleotide sequence analysis. Proc Natl Acad Sci U S A. 1979 May;76(5):2232–2235. doi: 10.1073/pnas.76.5.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizusawa S., Nishimura S., Seela F. Improvement of the dideoxy chain termination method of DNA sequencing by use of deoxy-7-deazaguanosine triphosphate in place of dGTP. Nucleic Acids Res. 1986 Feb 11;14(3):1319–1324. doi: 10.1093/nar/14.3.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn M. D., Thireos G., Greer H. Temporal analysis of general control of amino acid biosynthesis in Saccharomyces cerevisiae: role of positive regulatory genes in initiation and maintenance of mRNA derepression. Mol Cell Biol. 1984 Mar;4(3):520–528. doi: 10.1128/mcb.4.3.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret J. M., Chater K. F. Phage-mediated cloning of bldA, a region involved in Streptomyces coelicolor morphological development, and its analysis by genetic complementation. J Bacteriol. 1985 Sep;163(3):965–972. doi: 10.1128/jb.163.3.965-972.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins P. W., Trimble R. B., Wirth D. F., Hering C., Maley F., Maley G. F., Das R., Gibson B. W., Royal N., Biemann K. Primary structure of the Streptomyces enzyme endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1984 Jun 25;259(12):7577–7583. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thomas J. O., Kornberg R. D. The study of histone--histone associations by chemical cross-linking. Methods Cell Biol. 1978;18:429–440. [PubMed] [Google Scholar]
- Westpheling J., Ranes M., Losick R. RNA polymerase heterogeneity in Streptomyces coelicolor. Nature. 1985 Jan 3;313(5997):22–27. doi: 10.1038/313022a0. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]