Abstract
The adult Drosophila retina is a highly polarized epithelium derived from a precursor tissue that is initially symmetric across its dorsoventral axis. Specialized 90° rotational movements of subsets of cells, the ommatidial precursors, establish mirror symmetry in the retinal epithelium. Myosin II, or Zipper (Zip), a motor protein, regulates the rate at which ommatidia rotate: in zip mutants, the rate of rotation is significantly slowed. Zip is concentrated in the cells that we show to be at the likely interface between rotating and non-rotating cells: the boundary between differentiated and undifferentiated cells. Zip is also robust in newly added ommatidial cells, consistent with our model that the machinery that drives rotation should shift to newly recruited cells as they are added to the growing ommatidium. Finally, cell death genes and canonical Wnt signaling pathway members genetically modify the zip phenotype .
Keywords: Zipper, myosin II, ommatidial rotation, ommatidial orientation, Drosophila eye development, cell movements, cell death
Background
The movement and rearrangement of cells is essential for morphogenesis and development. Epithelial movement is coordinated among groups of cells in some instances, contributing to events such as gastrulation in vertebrates and dorsal closure in the Drosophila embryo (Djiane et al., 2000; Formstone and Mason, 2005; Kaltschmidt et al., 2002; Mlodzik, 2002; Morel and Arias, 2004; Tada et al., 2002; Winklbauer et al., 2001). In other events, cells move independently of their neighbors, such as during neural crest cell migration (Locascio and Nieto, 2001). In these cases, cells on the leading edge extend projections into the extra-cellular matrix and then contract the extensions to pull the cells forward. A distinct type of movement occurs when cells rearrange themselves within an epithelium, for example when cells intercalate between one another during convergent extension (CE) and when groups of cells move past one another, such as when the border cells of the ovary collectively migrate between the nurse cells (Montell, 2003). A similar cellular rearrangement takes place in the developing Drosophila eye when discrete groups of cells rotate on an axis within the epithelium. The mechanisms that produce the force for this latter type of movement are not understood.
In the developing Drosophila compound eye, 800 groups of cells undergo a 90° rotational movement within the epithelium. These groups of cells constitute the precursors to the ommatidia, the unit eyes of the compound eye (Fig. 1A). The direction in which the ommatidial precursors rotate is dictated by their location in the eye: those in the dorsal half rotate counterclockwise whereas those in the ventral half rotate clockwise (Fig. 1B, blue arrows). These rotational movements set up a dorsoventral midline, known as the equator (Fig. 1A: yellow line).
Figure 1. The Drosophila eye is a polarized epithelium.
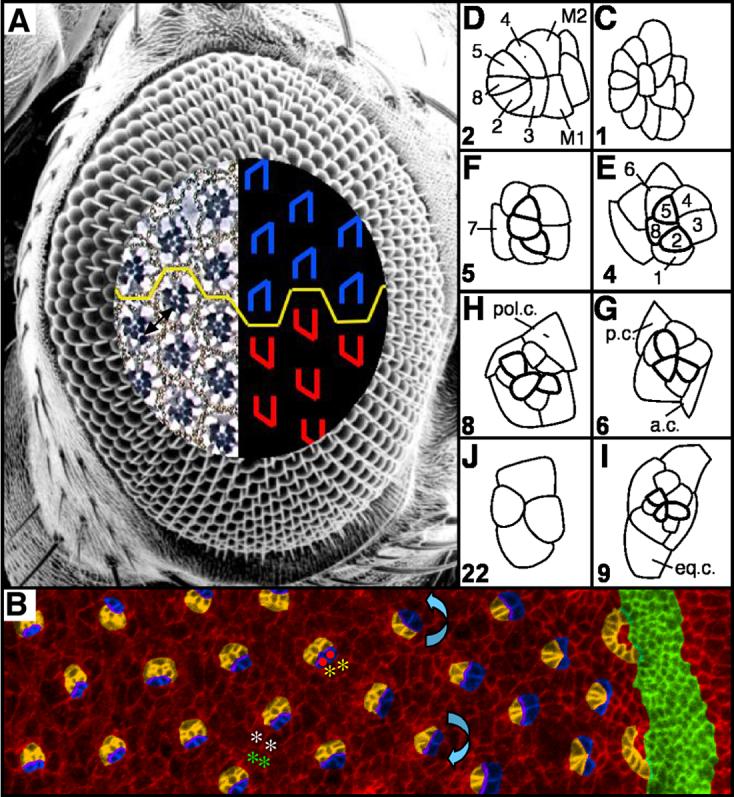
(A) Scanning electron micrograph of a Drosophila compound eye. A tangential demi-section reveals the underlying cellular morphology of the eye. The rhabdomeres, or light-sensitive organelles of the photoreceptors, form chiral trapezoids that are shown schematically in blue (dorsally-oriented trapezoids) and red (ventrally-oriented trapezoids). The trapezoids are oriented perpendicular to the equator, the line of mirror symmetry that divides the eye into dorsal and ventral halves (yellow line). Arrow highlights ommatidia separated by 4°. (B) Apical surface of a third instar eye imaginal disc immunostained with α-Arm and pseudocolored to highlight the morphogenetic furrow (green), arcs (orange), photoreceptors R8, R2, R5, R1, R7 and R6 (orange), and R3 and R4 (blue). The direction of rotation is indicated by blue arrows. The interface between rotating cells could reside between undifferentiated, interommatidial cells (white and green asterisks) or between rotating photoreceptors (red dots) and stationary undifferentiated cells (yellow asterisks). (C-J) Video lucida drawings of a maturational time line of ommatidial precursors. See text for details. Numbers in lower left hand corners indicate row number. M1, M2: mystery cells; 1-8: photoreceptor cells; a.c. anterior cone cell; p.c. posterior cone cell; pol.c.: polar cone cell; eq.c.: equatorial cone cell. Anterior to the right.
The rotating units of cells are partially assembled several hours before they begin to rotate. Ommatidial assembly and pattern formation in the compound eye proceed in a wave that moves from the posterior to the anterior of the imaginal disc. The leading edge of this wave is marked by a groove in the epithelium, the morphogenetic furrow (Ready et al., 1976). The first organized groups of cells to emerge from the furrow form an arc of equipotent cells (Dokucu et al., 1996; Wolff and Ready, 1991b). One cell from the arc differentiates to form the photoreceptor (R) 8 (R8) cell (Baker et al., 1996; Dokucu et al., 1996). R8’s two adjacent neighbors in the arc subsequently adopt the R2 and R5 fates, followed by R3 and R4, completing assembly of the ommatidial precluster (Tomlinson, 1985). All subsequent cells that are either incorporated into the ommatidium or killed off by cell death are generated in a wave of cell divisions known as the second mitotic wave (Campos-Ortega, 1977; Wolff and Ready, 1991b). R1 and R6 are the next two cells to be recruited into the ommatidium, followed by R7. The last class of cells to be added to the growing ommatidium during the larval phase of development are the non-neuronal lens-secreting cone cells; the anterior and posterior cone cells are added first, followed by the polar and equatorial cone cells (Fig. 1C-J; this figure was originally printed as Fig. 15 in “Pattern formation in the Drosophila retina,” by Wolff and Ready in “The Development of Drosophila melanogaster,” reprinted here with permission by Cold Spring Harbor Laboratory Press).
The mechanisms that drive the movement of these rotating groups of cells are unclear, although several genes have been implicated in regulating ommatidial rotation. These “rotation genes” differ from the class of tissue polarity genes in that they impact only the degree to which ommatidia rotate, but do not affect cell fate and chirality; the tissue polarity genes influence all three. Among these rotation-specific genes are members of the Jun kinase pathway, members of the EGF signaling pathway, and a serine/threonine kinase similar to the MEKs, nemo (nmo) (Brown and Freeman, 2003; Choi and Benzer, 1994; Chou and Chien, 2002; Freeman et al., 1992; Gaengel and Mlodzik, 2003; Lee et al., 1996; Mirkovic et al., 2002; Strutt and Strutt, 2003).
In an effort to uncover molecules likely to regulate this rotational movement, we conducted a dominant modifier screen in a nmo misexpression background. We identified the non-muscle myosin zipper (zip) as a suppressor of misexpressed nmo. Although we subsequently showed that zip and nmo are not bona fide genetic interactors, as loss-of-function zip does not interact with loss-of-function nmo, concurrent work revealed that zip does affect the rotation process.
Zip regulates several types of cellular movements, including dorsal closure, CE, and border cell migration (Bertet et al., 2004; Edwards and Kiehart, 1996; Franke et al., 2005; Mizuno et al., 2002). In these processes, Zip provides the force for movement by contracting a myosin network. During dorsal closure, Zip contracts a belt of actin like a purse string, in which each cell on the perimeter of the hole contributes to the “belt” (Franke et al., 2005). In CE, populations of cells intercalate with one other when Zip causes vertically-oriented actin fibers to contract (Zallen and Wieschaus, 2004). Zip provides the force for border cell migration during oocyte development by contracting actin filaments that extend between the border cells and nurse cells (Fulga and Rorth, 2002). Given its widespread roles in generating forces for movement, by analogy, zip may contribute to the force for ommatidial rotation.
Here, we show that Zip, a motor protein, is essential for ommatidial rotation. Zip accumulates in cells recruited into the growing ommatidium (photoreceptors and cone cells), cells that we show to be at the interface between rotating and non-rotating cells. This localization pattern is intriguing in that, in the simplest scenario, this interface represents the locale for the “machinery” that drives rotation. Consistent with the hypothesis that Zip is part of the rotation machinery, broadly defined, we show that, as new cells are added to the maturing ommatidial precursor and the interface between rotating and stationary cells consequently changes, Zip localization undergoes a parallel accumulation in the newly recruited (and newly rotating) cells. Finally, we show that wild type corrects the minor deviations in ommatidial orientation observed at the completion of rotation, probably through organizing the interommatidial lattice. Zip mutants fail to correct these deviations. Rotation-based patterning defects are likely primarily responsible for this failure, although genetic interaction studies suggest Zip may interact with canonical Wnt signaling and cell death to correct these deviations, as the ommatidial orientation phenotype of zip mutant ommatidia is enhanced by decreased activity of components of the canonical Wnt signaling pathway and suppressed by decreased activity of cell death genes. Notably, canonical Wnt signaling has been implicated in cell death (Ahmed et al., 1998; Cox et al., 2000; Freeman and Bienz, 2001). The suppression of the zip phenotype by cell death genes raises the possibility that zip regulates ommatidial orientation by cell death-dependent patterning of the pigment cell lattice.
Materials and Methods
Genetics
Fly strains used: Canton S, w1118, nmoP1/TM6, UAS>nmo, zip1/CyO, w;zip1, FRT42D/Cyo, zipEbr/CyO (gift from R. Ward (Ward et al., 2003)), cn1,zip02957/CyO, UAS>zipDN (gift from D. Kiehart), UAS>zip-GFP (gift from A. Brand (Barros et al., 2003)), sev>GAL4, GMR>GAL4, Drok2/FM7a, fzKD4A/CyO, dsh1, dsh3/FM7a, stbm6cn, fmiE59/CyO, pksple/CyO, arm8/FM7a, Df(H99)/TM6b (gift from R. Cagan), hid05014/TM6b (gift from R. Cagan), GMR>P35. ey>FLP was used to generate zip clones.
Immunohistology
Third instar larval eye imaginal discs and mid-pupal eyes (39 hours after puparium formation at 25°C) were dissected and processed as described (Wolff, 2000). Tissue was incubated in primary antibody overnight at 4°C at concentrations of 1:10 for α-Armadillo (α-Arm; mouse monoclonal, Developmental Studies Hybridoma Bank), 1:20 for phalloidin (Molecular Probes), and 1:500 for α-Zipper (rabbit polyclonal, generous gift from D. Kiehart (Kiehart and Feghali, 1986)). Antibodies conjugated to Alexafluor fluorescent dyes were used as secondary antibodies (Molecular Probes). Fluorescent images were collected using a Leica TCS SP2 confocal microscope.
Phenotypic analyses
Adult eyes were fixed, embedded and sectioned according to standard protocols (Wolff, 2000). Angles were measured between two vectors, one running parallel to the equator and the second drawn through the rhabdomeres of photoreceptors (R) R1, R2, and R3, using the ImageJ software. The number of ommatidia and eyes scored is as follows: Canton S: 2103 ommatidia from 11 eyes; UAS>zip;sev>Gal4/sev>Gal4: 1050 ommatidia from 8 eyes; UAS>zipDN;sev>Gal4: 723 ommatidia from 8 eyes. UAS>zipDN;sev>Gal4/nmoP1: 568 ommatidia from 6 eyes; UAS>zipDN/UAS>zip;sev>GAL4: 448 ommatidia from 5 eyes; UAS>zipDN/zip02957;sev>GAL4; 584 ommatidia from 6 eyes; GMR>P35: 690 ommatidia from 8 eyes.
sev>zipDN interactions were scored as described previously in the following number of ommatidia and eyes: fzKD4A, 938 ommatidia from 10 eyes; Drok2, 751 ommatidia from 8 eyes; dsh1, 534 ommatidia from 5 eyes; dsh3, 840 ommatidia from 8 eyes; stbm6cn, 1192 ommatidia from 12 eyes; fmiE59, 505 ommatida from 6 eyes; pksple, 774 ommatidia from 8 eyes; arm8, 609 ommatidia from 7 eyes; Df(H99), 881 ommatidia from 8 eyes; hid05014, 573 ommatidia from 6 eyes.
Third larval instar eye discs were dissected, fixed and immunolabeled with α-Arm, as described (Wolff, 2000). Rotation angles were measured between two vectors, one drawn parallel to the equator and the second running between the R3/4 interface and through the center of R8. ImageJ software was used to measure the angles. The number of ommatidia scored is as follows. w1118: row 2, 92 ommatidia; row 3, 101 ommatidia; row 4, 101 ommatidia; row 5, 101 ommatidia; row 6, 100 ommatidia; row 7, 100 ommatidia; row 8, 100 ommatidia; row 9, 99 ommatidia; row 10, 99 ommatidia; row 11, 99 ommatidia; row 12, 98 ommatidia; row 13, 98 ommatidia; row 14, 88 ommatidia; row 15, 80 ommatidia; from 12 eyes; UAS>zip; sev>GAL4: row 2, 54 ommatidia; row 3, 58 ommatidia; row 4, 57 ommatidia; row 5, 55 ommatidia; row 6, 53 ommatidia; row 7, 54 ommatidia; row 8, 54 ommatidia; row 9, 52 ommatidia; row 10, 49 ommatidia; row 11, 47 ommatidia; row 12, 42 ommatidia; row 13, 38 ommatidia; row 14, 33 ommatidia; row 15, 34 ommatidia; from 5 eyes; UAS>zipDN;sev>GAL4: row 2, 43 ommatidia; row 3, 68 ommatidia; row 4, 76 ommatidia; row 5, 72 ommatidia; row 6, 76 ommatidia; row 7, 74 ommatidia; row 8, 74 ommatidia; row 9, 75 ommatidia; row 10, 70 ommatidia; row 11, 64 ommatidia; row 12, 66 ommatidia; row 13, 64 ommatidia; row 14, 60 ommatidia; row 15, 57 ommatidia; from 7 eyes; UAS>zipDN;GMR>GAL4; row 2, 106 ommatidia; row 3, 110 ommatidia; row 4, 103 ommatidia; row 5, 102 ommatidia; row 6, 108 ommatidia; row 7, 103 ommatidia; row 8, 96 ommatidia; row 9, 81 ommatidia; row 10, 80 ommatidia; row 11, 63 ommatidia; row 12, 68 ommatidia; row 13, 52 ommatidia, row 14, 50 ommatidia; row 15, 46 ommatidia; from 14 eyes; GMR>P35: row 2, 56 ommatidia; row 3, 59 ommatidia; row 4, 60 ommatidia; row 5, 64 ommatidia; row 6, 63 ommatidia; row 7, 62 ommatidia; row 8, 63 ommatidia; row 9, 61 ommatidia; row 10, 59 ommatidia; row 11, 57 ommatidia; row 12, 49 ommatidia; row 13, 52 ommatidia; row 14, 53 ommatidia; row 15, 54 ommatidia; from 7 eyes.
The data illustrated in Fig. 4C was obtained from α-Arm-stained eye discs; angles were measured as described above. The number of ommatidia scored for each developmental stage is as follows: two mystery cells, 53 ommatidia; one mystery cell, 40 ommatidia; 5-cell stage, 68 ommatidia; 7-cell stage, 99 ommatidia; 8 cell stage, 112 ommatidia; posterior and anterior cone cell stage, 151 ommatidia; polar cone cell stage, 122 ommatidia; equatorial cone cell stage, 92 ommatidia.
Figure 4. The interface for rotation resides between the photoreceptors/cone cells and undifferentiated cells.
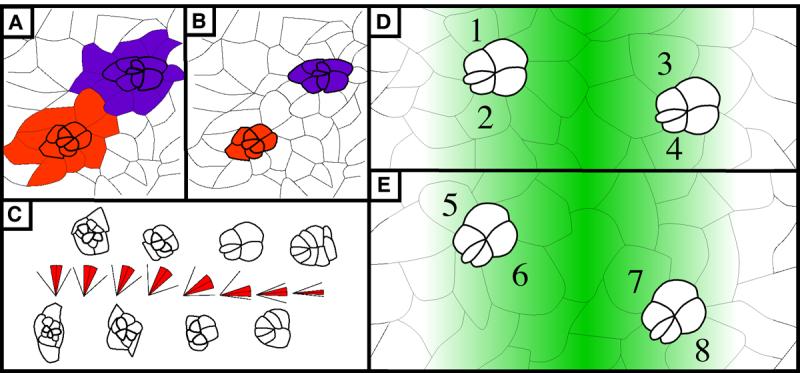
(A) and (B): Two models illustrating potential interfaces between rotating cells. In (A) like-colored cells are linked together and move as a group so the interface is at the purple/orange border. In (B), undifferentiated cells do not rotate with differentiated cells. The interface between rotating and stationary cells is between the colored and uncolored cells. (C) Average angle of orientation by developmental stage. Each “stick diagram” corresponds to the ommatidial form that lies directly above or below the diagram. In the diagrams, the center line indicates the average angle, red shading highlights standard deviation, and outermost lines indicate minimum (lower line) and maximum (higher line) degree of rotation seen for each developmental stage indicated. (D) and (E): Schematic representation of relative likelihood of anterior and posterior cone cells being recruited from the BUdR-labeled pool of cells. Green-colored gradient represents probability of a cell being labeled with BUdR. In both panels, two ommatidial precursors are shown, one anterior to and the second posterior to, the BUdR-labeled band. Darker cells are more likely to be labeled since they lie closer to this band. Numbers identify cells that will become anterior and posterior cone cells. In (D), cells are recruited prior to rotation (as modeled in Fig. 4B), so cells numbered 1 through 4, which have equivalent levels of shading, share equal probabilities of being labeled. When examined as pupal eyes, equal numbers of labeled anterior and posterior cells are predicted. In (E), cells are recruited after rotation begins (as modeled in Fig. 4C). Consequently, two cells are recruited from the pool that is more likely to be labeled (6,7, dark green) whereas the other two cells are recruited from the pool that is less likely to be labeled (5,8, pale green). When examined as pupal eyes, the number of labeled anterior and posterior cells is predicted to be disproportionate. See text for details. Anterior is to the right for all panels.
Results
zip exhibits an ommatidial orientation phenotype in adult eyes
zip was identified in a screen for modifiers of the rotation phenotype of the serine threonine kinase, nemo (Choi and Benzer, 1994). zip dominantly suppresses the sev>nmo rotation phenotype from 44 +/- 16 to 15 +/- 4 percent errors (p<2×10-10). While loss-of-function genetic assays suggest zip and nmo do not interact (data not shown), we cannot rule out the possibility that the absence of an interaction may be a consequence of the alleles used in the assay. Concurrent work revealed zip function is essential for normal ommatidial orientation.
Homozygous zip1 (a null allele) flies die prior to ommatidial rotation. Clones of zip1 tissue could not be analyzed since GFP-negative tissue bordering the twin spots in eye imaginal discs is α-Arm negative, and in adults, only small scars are apparent bordering the twin spots (data not shown). Genotypically wild-type ommatidia adjacent to these scars exhibit orientation errors (Fig. 2A). These orientation defects are likely due to zip-induced flaws in the pigment cell lattice (see below).
Figure 2. zip exhibits an orientation phenotype.
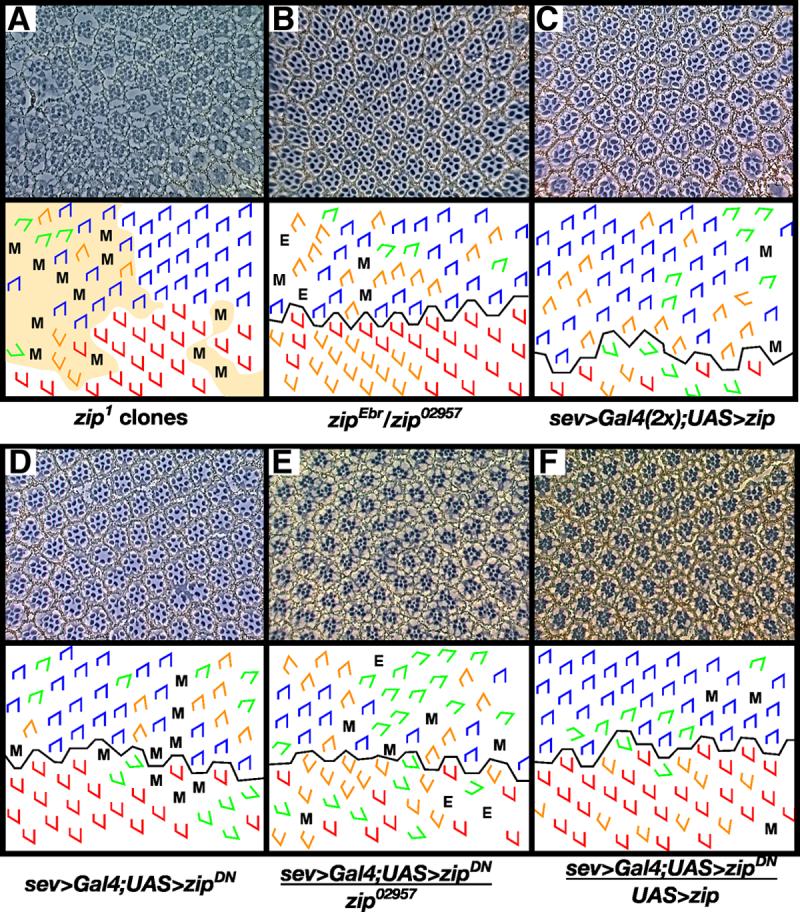
Clones of the null allele zip1 (A) and hypomorphic transheterozygous flies (zipEbr/zip02957) (B) exhibit rotation errors. In (A), shaded area denotes regions of phenotypically mutant tissue, presumably a consequence of loss of zip mutant cells since no w- photoreceptors can be identified. Misexpression of zip (C) and zipDN (D) under the sev promoter results in defects in ommatidial rotation. To demonstrate the efficacy of the sev>zipDN transgene, zip dosage was decreased (E) or increased (F) in a sev>zipDN background. Decreasing zip dosage using zip02957 enhances the sev>zipDN phenotype (E) whereas increasing zip dosage using a second copy of the UAS>zip transgene suppresses the sev>zipDN phenotype. M, missing photoreceptors; E, extra photoreceptors; blue trapezoids: dorsal ommatidia; red trapezoids: ventral ommatidia; green trapezoids: under-rotated ommatidia; orange trapezoids: over-rotated ommatidia.
Since strong loss-of-function alleles of zip severely disrupt retinal morphology (Baumann, 2004) and thereby preclude analysis of a potential rotation phenotype, we instead attempted to enable initial growth and recruitment of photoreceptors to take place normally by removing less Zip activity. We partially removed zip function using either a viable, heteroallelic fly (zipEbr/zip02957) (Fig. 2B) or a dominant negative transgene, zipDN (Fig. 2D). zipDN transgenic flies mimic loss-of-function alleles (Burns et al., 1995; Franke et al., 2005). The transgene was expressed using the eye-specific driver sev>Gal4, which drives expression in photoreceptors R3, R4, R7 and the cone cells. To test the efficacy of this transgene in the eye, we modified Zip levels and assayed for the predicted interactions. Decreasing endogenous Zip activity in the sev>zipDN background enhances the sev>zipDN phenotype, whereas increasing Zip activity suppresses the phenotype (Fig. 2D, E, Table 1), indicating that ZipDN effectively blocks endogenous Zip activity in the eye.
Table 1.
Average angle of orientation in adult eyes.
| Genotype | Angle of orientation (+/- SD) | N | |
|---|---|---|---|
| w1118 | 92 (+/- 4) | 11 | |
| zipEbr/zip02957 | 99 (+/- 17) | 8 | |
| sev>Gal4/sev>Gal4;UAS>zip | 95 (+/- 27) | 8 | |
| sev>Gal4;UAS>zipDN | 85 (+/- 20) | 8 | |
| GMR>P35 | 91 (+/- 12) | 8 | |
| sev>Gal4;UAS>zipDN /* | |||
| zip02957 | 89 (+/- 28) † | 6 | |
| UAS>zip | 88 (+/- 15) † | 5 | |
| Drok2 | 82 (+/- 23) † | 8 | |
| fmiE59 | 85 (+/- 21) | 6 | |
| strabismus6cn | 86 (+/- 21) | 12 | |
| pricklesple | 87 (+/- 18) | 8 | |
| dishevelled1 | 88 (+/- 15) † | 5 | |
| dishevelled3 | 91 (+/- 25) ‡ | 8 | |
| frizzledKD4A | 79 (+/- 34) ‡ | 10 | |
| armadillo8 | 78 (+/- 37) ‡ | 7 | |
| hid05014 | 87 (+/- 14) | 6 | |
| Df(3L) H99 | 90 (+/- 11) ‡ | 8 |
N = number of eyes scored.
Statistical significance measured by Student’s t-test
p-value < .01
p-value < .001
Ommatidia in wild-type eyes appear perfectly aligned with one another. There is, however, a small amount of variation in ommatidial orientation, by our measurements (92° +/-4; Fig. 1A: arrow). This variation is likely due to the combined effect of many factors, including curvature of the eye (which leads to slight skewing of at least some ommatidia in sections), minor morphological inconsistencies in ommatidia (i.e. rhabdomere shape, alignment), and possibly even imperceptible inherent variation in orientation (by our assessments, it is not possible to detect 5° of variation without the use of a protractor).
Ommatidia in zipEbr/zip02957 adult eyes exhibit a presumptive “rotation” phenotype in which ommatidia appear to both over- and under-rotate, although more ommatidia over-rotate (99° +/- 17) (Fig. 2B, Table 1). (Deviation from the wild type 90° is generally assumed to result from defects in rotation. Our use of “presumptive” is intended to caution that additional factors could, and in fact do (discussed below), contribute to this phenotype. For simplicity, we do not use the word “presumptive” throughout this discussion.) Flies expressing the dominant negative transgene also exhibit both over- and under-rotation defects, producing an average ommatidial angle of 85° +/- 20 (Fig. 2D, Table 1). In contrast, misexpression of zip+ generates a phenotype in which more ommatidia over-rotate than under-rotate (95° +/- 27) (Fig. 2C, Table 1). The large standard deviation indicates these phenotypes are significant, even though the average angle of rotation is not dramatically changed. The mild disruption of the average angle may be a consequence of compromising Zip function in only a fraction of cells in each ommatidium.
zip regulates the rate of ommatidial rotation
One caveat to scoring a presumptive rotation phenotype in adult eyes is that patterning events that take place following the completion of rotation could affect ommatidial packing and could thereby affect ommatidial orientation. Such secondary “orientation-based” defects would therefore be mistaken for rotation-based defects. To rule out the possibility that post-rotation patterning events contribute to the zip phenotype, and to confirm that zip does play a role in ommatidial rotation, both the degree to which ommatidial precursors rotate and the rate at which they rotate were measured in third instar eye imaginal discs. These parameters were measured in wild type, sev>zipDN, GMR>zipDN and sev>zip+ eye discs (Fig. 3).
Figure 3. zip regulates the rate of ommatidial rotation.
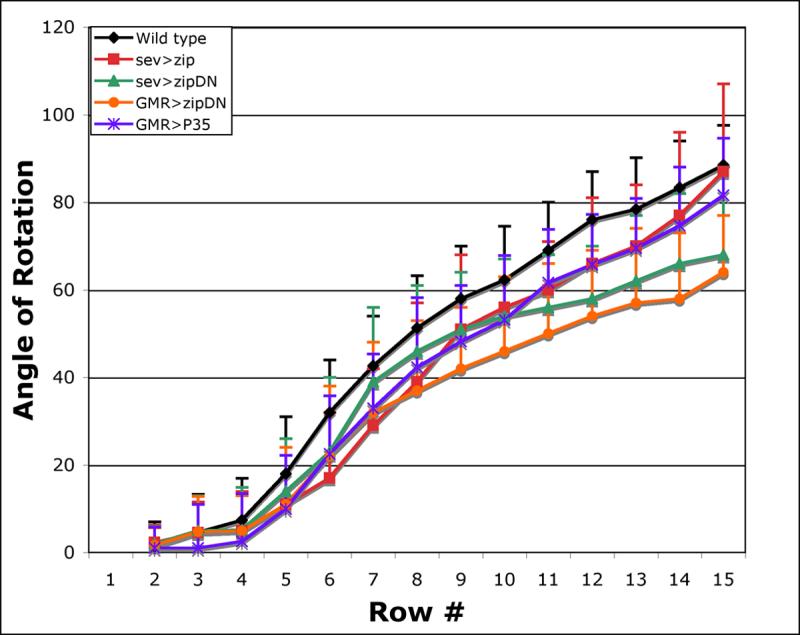
Wild-type rotation begins at row 4 and is effectively complete by row 15 (black). Decreasing zip function using the sev and GMR promoters to drive zipDN slows the rate of rotation (sev>zipDN, green, and GMR>zipDN, orange). Misexpressing wild-type zip in a subset of cells also disrupts the rate of rotation (sev>zip, red). Misexpressing P35 under the GMR promoter delays rotation in a pattern similar to sev>zip (purple). Wild-type is statistically different from: sev>zip in rows 6-8 and 11-12 (t-test, p<.001); sev>zipDN in rows 6 and 9-15 (t-test, p<.001); GMR>zipDN in rows 5-15 (t-test, p<.001); GMR>P35 in rows 5-14 (t-test, p<.001).
Angles were defined by 1) a vector drawn between the R3/R4 interface through the center of R8 and 2) a line running parallel to the equator (Fig. 3). Row number was assigned based on the morphology of the ommatidium closest to the furrow in a given column (a column comprises a line of ommatidia arranged anterior to posterior; row numbers as defined by Wolff and Ready, (Wolff and Ready, 1993)). For example, if the “youngest” ommatidial precursor in a column was an arc, that row was assigned a classification of row 1; its adjacent, posterior neighbor was assigned row 2, etc.. The values for each row were averaged to derive an average angle of orientation for the respective rows.
In wild-type eye discs, rotation begins at row four and essentially reaches completion at row 15 (88° +/- 9°). Although the average angle of orientation is virtually 90° by row 15, many clusters have either not yet reached 90° or they have rotated beyond the 90° stopping point (standard deviation: +/- 9°). The significant standard deviation in ommatidial orientation at row 15 indicates that the mechanisms that drive ommatidial rotation are not tuned to stop precisely at 90°; rather, some later patterning event precisely aligns the lattice once rotation is complete. These observations support the hypothesis of the “locking mechanism” initially proposed by Brown and Freeman (2003) to explain the presence of a phenotype in EGFR pathway mutant adults in the absence of a larval phenotype.
In addition, in wild-type eye discs, rotation is initially fast, rotating at a rate of ∼10-15° per row between rows four and six, and then slows to a rate of ∼5-10° per row. The fast phase continues until the point at which the cone cells are being added whereas the slower phase carries them the rest of the way. These two distinct rates likely underlie the apparent pause in rotation reported by Gaengel et al. (Gaengel and Mlodzik, 2003), however, it is important to note that we do not see a pause.
Loss of zip function, using sev and GMR to drive a dominant negative zip transgene, slows the rate of ommatidial rotation (Fig. 3). By row 15, when rotation is essentially complete in wild type, ommatidial precursors have rotated just 64° +/-13 in GMR>zipDN eyes and 68° +/-20 in sev>zipDN eyes. The rate of rotation in zipDN eyes driven by GMR is significantly different from the wild-type rate from the onset of rotation whereas it generally takes longer for sev to exert its effects (Fig.3). We attribute the eventual slowdown in sev>zipDN eyes to the recruitment of new, sev-positive cells into the ommatidium. R7 is recruited into the assembling ommatidial precursor between rows 5 and 6 and the anterior and posterior cone cells are added between rows 6 and 7. As these new cells are recruited, overall levels of ZipDN increase, exerting a stronger effect on the rate of rotation and leading to a slowdown beginning at row 7 in sev>zipDN eyes (Fig. 3).
There is a dramatic discrepancy between the average angle of orientation in sev>zipDN mutants in eye imaginal discs relative to the adult. By row 15 - a point that marks the end of ommatidial rotation in wild-type discs - zip ommatidial precursors have rotated 68° (+/-20), whereas in the adult, they have rotated an additional 20° (85° +/- 20) (Fig. 3). Two plausible mechanisms that could account for this additional movement are prolonged rotation or normal patterning events that occur during pupal life (discussed below). The notion that normal patterning events could turn ommatidia an additional 20° is unlikely, given these events do not have a similar effect on wild-type development. We therefore favor a model in which rotation continues beyond the normal stopping point.
Misexpression of wild-type zip might be predicted to lead to an increased rate of rotation based on the loss-of-function phenotype, however, it leads to an initial, modest delay in rotation, a phenomenon we cannot explain. Following this initial delay, rotation proceeds at the same rate as wild type. By row 15, the average angle of rotation in sev>zip+ eye discs is virtually equal to that of wild type, yet the standard deviation is almost double that of wild type. The large standard deviation is a reflection of the fact that far more ommatidia both fall short of and surpass the 90° stopping point, perhaps as a consequence of interference with the rotation machinery by excess zip activity. These combined results implicate zip in a role for regulating the speed at which ommatidia rotate.
Cone cells become morphologically distinct starting at 30 - 45° of rotation
Zip, a motor protein that contributes to the regulation of rotation, could participate directly or indirectly in the generation of force for rotation. To devise a model for how zip may act in this capacity, it was necessary to first identify those cells between which the force needs to be generated. In the simplest scenario, the force is likely to be generated at the interface between moving and stationary cells.
It has long been known that the photoreceptor clusters are members of the moving subset of cells, but whether the future cone cells and all, some, or none of the “late-differentiating” interommatidial cells (the future pigment cells) are also members of this moving cohort has not been defined. Intercellular contacts could be remodeled to enable ommatidial precursors to rotate within the epithelium in one of two places: 1) between undifferentiated interommatidial cells (the future pigment and cone cells; green and white asterisks, Fig. 1B; Fig. 4A, and see below for details), in which case those interommatidial cells contacting one ommatidial precursor would rotate with that precursor while the adjacent interommatidial cells contacting the neighboring ommatidial precursor would rotate with the neighboring ommatidial precursor (Fig. 4A) or 2) between the photoreceptor cohort (red dots, Fig. 1B) and their neighboring, undifferentiated, interommatidial cells (yellow asterisks, Fig. 1B; Fig. 4B).
As a first step in interpreting labeling studies designed to address this question (described below), the time of cone cell recruitment was correlated with the degree to which ommatidial precursors had rotated (Fig. 4C). The unit eyes of the compound eye are built by successive recruitment of cells from the surrounding pool of undifferentiated cells. Those undifferentiated cells that make the right set of contacts and receive the appropriate signals will differentiate to adopt the fates occupied by a given niche. Photoreceptors R1 and R6, followed by R7, are recruited first (Ready et al., 1976; Wolff and Ready, 1993). The anterior and posterior cone cells are recruited next, followed a short time later by the polar and, finally, equatorial cone cells (Wolff and Ready, 1993). Once this core is compiled, the pigment and bristle cells are added during pupal life to complete the ommatidium.
Eye imaginal discs were examined at two focal planes to determine when during rotation the cone cells are recruited. Third instar eye discs were labeled with α-Armadillo (α -Arm) to label cell outlines at the level of the adherens junctions and were compared to more apical outlines of cells, as revealed by lead sulfide. The cone cell precursors establish stereotypic contacts with the photoreceptors; these contacts were used as a defining feature to identify the cone cells. Observations of these discs indicate that rotation becomes apparent when just five cells comprise the rotating group: the future photoreceptors R8, R2, R5, R3 and R4. Comparisons of the two preparations revealed that contacts and stereotypic cellular morphologies are established apically and consolidated basally. The timeline described here is derived from images using the adherens junction marker, so use of an alternative, more apical marker is expected to give subtly different results.
By the time the ommatidial precursors rotate approximately 65° (+/- 12), the anterior and posterior cone cells are unambiguous members of the ommatidial complex, as defined by α-Arm (Fig. 4C). By this point the cone cells have not yet adopted their final morphology, but they do occupy their ultimate positions in the ommatidium. In many cases, by roughly 55° (+/- 13) of rotation, single cells occupy the sites of the future anterior and posterior cone cells; these cells likely will become the anterior and posterior cone cells (Fig. 4 D, E). The polar cone cell is clearly in position (and again stains robustly with α-Arm) by the time the ommatidial precursor has rotated 77° (+/- 13). The equatorial cone cell is not unambiguously positioned until the end of rotation (89° +/- 11). While these results do not establish if the precursors to the cone cells are carried with the photoreceptors from the commencement of rotation, as undifferentiated cells, they do establish that the anterior and posterior cone cells are recruited into the cluster and begin to rotate with the photoreceptors by 65° of rotation, at the very latest, the polar cone cell by 77°, at the latest, and that the equatorial cone cell is added essentially at the completion of rotation.
Ommatidial precursors move independently of their undifferentiated neighbors
To determine whether all, some or none of the interommatidial, undifferentiated cells (future cone and pigment cells) move with the photoreceptors, and thereby infer the location of the interface between rotating and stationary cells, a subset of undifferentiated cells was labeled prior to both rotation and differentiation. The ultimate fates and locations of these cells were then mapped in the fully patterned, mid-pupal epithelium, a method that enabled us to reconstruct which cells move during patterning. Third instar larvae were injected with a pulse of bromouridine deoxyri bose (BUdR) to label S phase cells that were dividing in the second mitotic wave, a narrow band of divisions that takes place between two successive rows of ommatidial precursors (Fig. 4 D,E). The following observations were made on data published by Wolff and Ready, 1991 (Fig. 7, Wolff and Ready, 1991, and reprinted here as Fig. 5; refer to Wolff and Ready, 1991 for methods and uncropped version of figure).
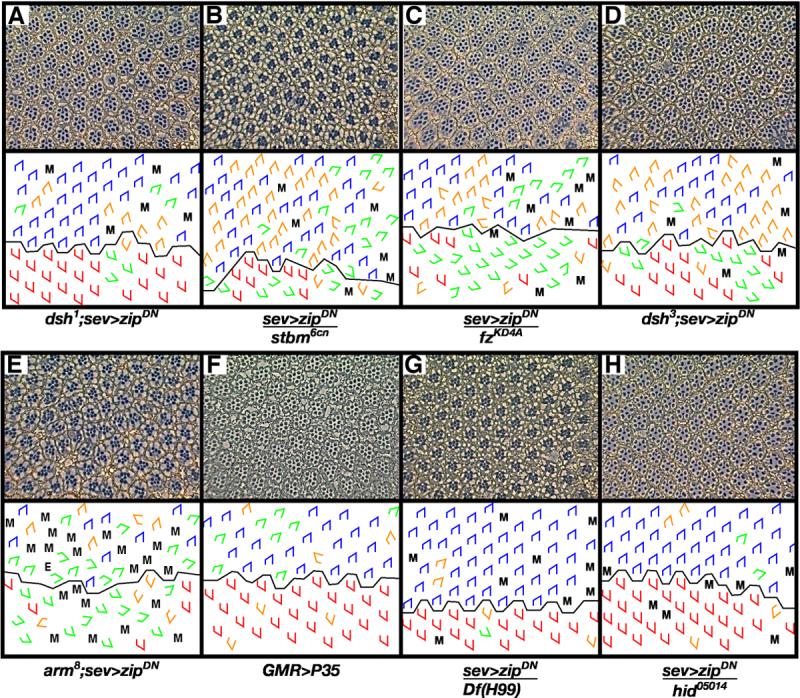
Figure 7. sev>zipDN interacts with canonical Wnt signaling and cell death genes.
sev>zipDN is dominantly suppressed by dsh1 (A), but no interaction is seen with the tissue polarity gene stbm6cn (B). However sev>zipDN is enhanced by fzKD4A (C) and dsh3 (D) (compare to figure 2D). An allele of arm (arm8) (E) specific for canonical Wnt signaling also enhances sev>zipDN. GMR>P35 (F) produces an ommatidial orientation phenotype, and a deficiency that removes grim, rpr, hid (Df (H99)) (G) and hid05014 (H) suppress sev>zipDN. M, missing photoreceptors; E, extra photoreceptors; blue trapezoids: dorsal ommatidia; red trapezoids: ventral ommatidia; green trapezoids: under-rotated ommatidia; orange trapezoids: over-rotated ommatidia.
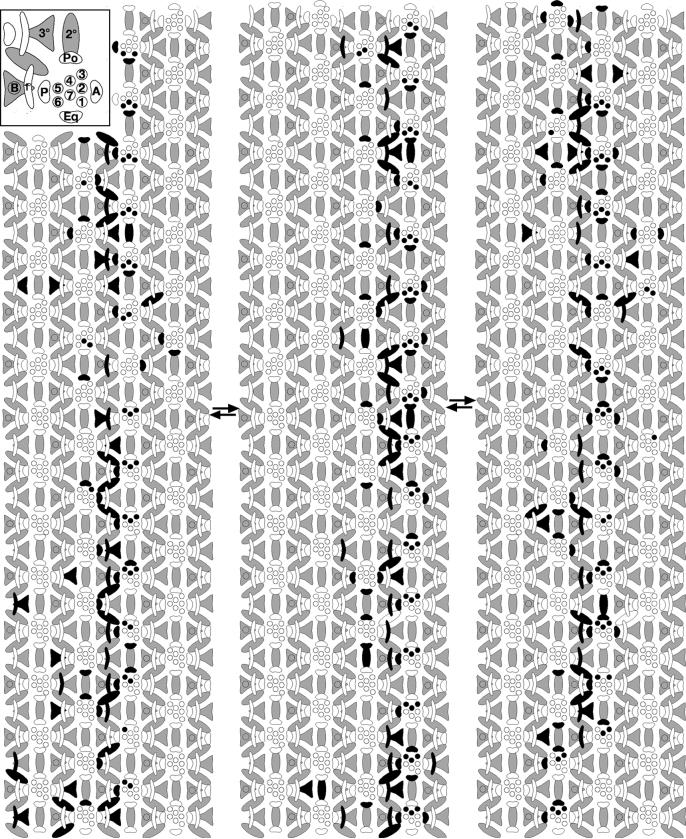
Figure 5. Positions and fates adopted by cells in second mitotic wave.
Each of the three schematics represents an independent eye. Black cells denote cells in S phase at the time a pulse of BUdR was administered. Whereas the anterior and posterior cone cells rotate with the photoreceptors, the future pigment cells - the undifferentiated, interommatidial cells of the third instar - retain their positions (i.e. they do not rotate with the photoreceptors). The interface between rotating and non-rotating cells therefore lies between the photoreceptors/cone cells and the interommatidial cells. See text for details and inset for cell types. 1-6: photoreceptor cells; B: bristle; 1°, 2° and 3°: primary, secondary and tertiary pigment cells, respectively; Po: polar cone cell; Eq: equatorial cone cell; A: anterior cone cell; P: posterior cone cell. Arrows denote the position of the equator. Anterior is to the right.
All cells that are not recruited into the preclusters (the preclusters consist of photoreceptor cells R8, R2, R5, R3, and R4) ultimately undergo a division in the second mitotic wave to produce the population of cells from which all subsequent cells are derived (Campos-Ortega, 1977; Ready et al., 1976; Wolff and Ready, 1991b). The pulse of BUdR therefore labels most cells between two ommatidial rows, cells that will later be recruited as R1, R6, R7, the four cone cells, the 1°, 2° and 3° pigment cells, and the bristle cells. (Note that contrary to what is suggested by the schematic (Fig. 4 D,E), since cell division is not synchronized in cells between successive rows, not every cell will be labeled with a single pulse of BUdR.) The final locations of cells recruited from the pool of labeled cells were evaluated at the completion of pattern formation. The phenomenon of ommatidial rotation-induced equatorial movement of cells recruited from the labeled band, as predicted by Tomlinson and Ready (Tomlinson et al., 1987) and as reported previously by Wolff and Ready (Wolff and Ready, 1991b), is most evident in the pattern of labeled photoreceptors R1, R6 and R7 (Fig. 5, Table 2). The labeled R1, R6 and R7 illustrated in Fig. 5 were recruited from the pool of labeled cells positioned posterior to the ommatidial precursor, as evidence by the observation that virtually all labeled photoreceptor R1, R6 and R7 cells lie in the row anterior to, rather than posterior to, the black band of label (72% and 9% respectively; Fig 5, Table 2).
Table 2.
Proportion of cell types labled with BUdR anterior and posterior to the labled pool of cells.
| Cell type | Row anterior to labeled pool | Row posterior to labeled pool |
|---|---|---|
| Photoreceptors (R1, R6, R7) | 72% | 9% |
| Polar cone cell | 7% | 40% |
| Equatorial cone cell | 48% | 20% |
| Anterior cone cell | 20% | 50% |
| Posterior cone cell | 60% | 20% |
Percents represent percentage of cells labeled with BUdR
There are two possible scenarios to describe which populations of cells rotate, (schematized in Fig. 4 A, B). In one scenario, the undifferentiated cells establish contacts with the photoreceptors prior to rotation. These contacts are fixed, such that cells that touch the photoreceptors move with the photoreceptors (Fig. 4A). In the second scenario, contacts between the photoreceptors and undifferentiated cells are remodeled over time, and only those cells with “established” fates (i.e. those that get recruited into the growing ommatidium) move with the ommatidial precursors (Fig. 4B). If the first scenario is correct, then the prospective anterior and posterior cone cells have equal access to the labeled pool of cells (Fig. 4D, cells 1-4), and an equivalent number of anterior and posterior cone cell precursors should be recruited from the labeled pool. If scenario two applies, there should be a bias in the ratio of labeled anterior to posterior cells because access to the pool of labeled cells becomes reciprocal for these two populations of cells: the anterior ommatidial precursor would have rotated such that the anterior cone cells would be recruited from a pool that lies further from the labeled band (Fig. 4E, cell 8) whereas the posterior cone cell from the same precursor would have greater access to the labeled pool (Fig. 4E, cell 7). Similarly, the anterior cone cell from the ommatidial precursor bordering the posterior edge of the band (Fig. 4E, cell 6) would have better access to the labeled cells than would its posterior partner (Fig. 4E, cell 5).
A comparison of numbers of labeled anterior cone cells from complementary faces of successive rows of ommatidia (i.e. both anterior and posterior to the labeled band (Fig. 4E, cells 6 and 8)), and of labeled posterior cone cells, again, from complementary faces of successive rows of ommatidia (Fig. 4E, cells 5 and 7), supports the second scenario: cells do not rotate with the photoreceptors until they have been recruited into their specific niches (Table 2). 19% of anterior cone cells from the anterior row are labeled compared to 59% of posterior cone cells from the same row, and 51% of anterior cone cells are labeled in the posterior row relative to just 15% in the corresponding cells (i.e. the posterior cone cells from the posterior row). An equivalent analysis for the polar and equatorial cone cells holds true, although the differences are less dramatic given the later recruitment of these cell types (Table 2). Similarly, analysis of the late-differentiating cells that adopt pigment cell fates also validates this model. In the case of the pigment cells, the tight, linear band of labeled 1°, 2° and 3° cells is a straightforward visual and qualitative representation of the data shown in Table 2 in that if these cells were carried by ommatidial rotation as described for scenario one, the band would be diffuse. Rather, the observation that the band is linear indicates that the cells do not get pulled along by the rotating photoreceptors.
The BUdR labeling studies, in conjunction with the description of cone cell recruitment with respect to degree of ommatidial rotation, indicate that the photoreceptors move independently of the pool of interommatidial cells, and that as cells are recruited into the growing ommatidium, they move with the photoreceptors. The interface between rotating and stationary cells is therefore initially between the photoreceptors and the undifferentiated interommatidial cells and later between the photoreceptors/cone cells and late-differentiating interommatidial cells.
Zip localizes to the apical domain in rotating ommatidia
Zip generates the driving force for diverse cellular movements. For example, the observation that Zip accumulates at the leading edge of the epithelium during dorsal closure led to the proposal that Zip produces a force similar to a purse string closing a hole (Franke et al., 2005). Zip has also been implicated in providing the force for cytokinesis since it accumulates in a ring at the cleavage furrow (Field and Alberts, 1995). Finally, during germ band extension, Zip accumulates at the longitudinal, but not the horizontal, edge of cells, leading to the model that Zip produces a force along the longitudinal membranes that drives their intercalation between neighboring cells (Bertet et al., 2004). To investigate the possibility that Zip may also provide the force for ommatidial rotation, we examined its subcellular distribution in rotating ommatidia.
Zip is strongly localized between rotating and non-rotating cells, as visualized by α-Zip, consistent with a role in providing the force for ommatidial rotation (Fig. 6A-A’). Furthermore, while Zip localization is uniform in undifferentiated cells, its expression in photoreceptor precursors is dynamic and asymmetric, as follows. In the arc stage of development (rows 1 and 2), Zip accumulates at the posterior membrane of arc cells (Fig. 6B,B’) in a fashion that resembles Zip localization during dorsal closure. When R7 and the anterior and posterior cone cells are recruited into the growing ommatidium, they become the new interface between rotating and stationary cells. As they adopt this new position, two events occur simultaneously. First, Zip localization becomes more prominent in these three cells (arrowhead, Fig. 6C’, C”, E’, E”), accumulating at the boundaries with the interommatidial cells. Second, Zip in R8, R2 and R5 appears to “retract” from their now-occluded interface, forming ring-like structures (arrow, Fig. 6E’,E”). This shift in Zip localization is consistent with our prediction that the machinery for rotation must shift to the outermost cells of the rotating unit.
Figure 6. Zip localizes strongly to differentiating cells.
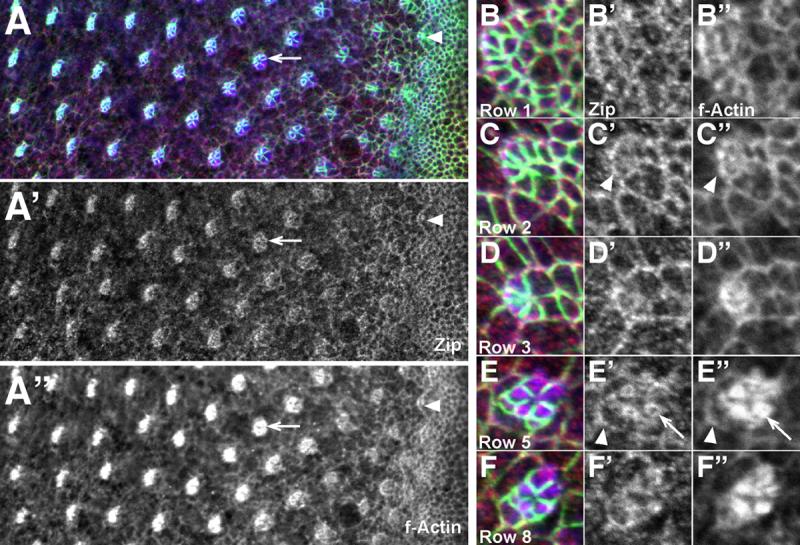
(A-F) Third larval instar eye disc stained with α-Arm (green), α-Zip (red), and phalloidin (blue). Co-localization of Zip (A’-F’) and actin (A”-F”) is evident in ommatidia before (arrowhead, A-A”) and during (arrow, A-A”) rotation. Close-ups of ommatidia (B-F) show dynamic co-localization of both actin and Zip. Accumulations are seen at the interface between ommatidial and undifferentiated cells (arrowheads C’, C”, E’, E”) and in the cytoplasm of rotating photoreceptors (arrows, E’ and E”). Anterior is to the right.
Zip also accumulates in cells that are not rotating, for example, cells in the morphogenetic furrow and in the arcs. Since Zip is used in a variety of cellular processes, it is not remarkable that these accumulations exist. One plausible role for Zip is that it regulates changes in cellular morphology in these cells (i.e. apical constriction of cells in the morphogenetic furrow and the changes in cell shape that are associated with closing the arc).
Since Zip tracks along filamentous actin (F actin), we examined F actin during ommatidial rotation and found that actin accumulation is coincident with Zip accumulation (Fig. 6A”-F”). Depending on whether the actin network is polarized or not, two types of forces could be generated by cooperation between actin and Zip. First, if the actin network is not polarized, a bidirectional contractile force could be generated (Kuczmarski et al., 1991). Alternatively, if the actin is bundled into a polarized network, Zip could generate a unidirectional force by moving along these bundles (Ishikawa et al., 2003). The coincident accumulation of actin and Zip specifically in rotating, and not non-rotating, cells is consistent with a role for Zip in generating a force, although Zip’s role in ommatidial rotation could certainly be more passive (see Discussion).
The absence of a genetic interaction between zip and nmo suggests Zip localization is unlikely to be altered in nmo mutant clones. Indeed, Zip is unaffected in nmo clones (data not shown). Both genes clearly affect rotation, but the genetic and localization data suggest their activities likely influence different genetic pathways.
zip interacts with canonical Wnt signaling pathway members
The Rho family GTPase member, Rho A, and its target effector, Drosophila Rho-associated kinase (Drok), are known regulators of the actin cytoskeleton (Winter et al., 2001). Mutations in these genes produce tissue polarity and rotation phenotypes in the eye and wing (Fanto et al., 2000; Verdier et al., 2006; Winter et al., 2001). These genes have been linked to tissue polarity signaling through genetic interactions with the tissue polarity-specific allele of disheveled (dsh), dsh1. Drok has also been linked to zip: they interact genetically (in the eye, zip suppresses the rough eye phenotype that results from overexpression of a constitutively active form of Drok (Verdier et al., 2006)) and Drok targets genes that regulate Zip (Winter et al., 2001). These results have led to the proposal that zip interacts with the tissue polarity genes to regulate tissue polarity in the eye (Fanto et al., 2000; Winter et al., 2001), although no direct evidence exists. We therefore tested for genetic interactions between sev>zipDN and Drok and sev>zipDN and members of the tissue polarity pathway. Drok mildly enhances the sev>zipDN phenotype (+/- 20 vs. +/-23; p<9×10-5), consistent with published results. Although dsh1 suppresses sev>zipDN (Fig. 7A, Table 2), neither flamingoE59 (fmiE59), pricklesple (pksple), nor strabismus6cn (stbm6cn) modify sev>zipDN (Fig.7B, Table 1). While the absence of interactions with three of the tissue polarity genes casts some doubt on the theory that zip interacts with the tissue polarity pathway (Verdier et al., 2006; Winter et al., 2001), it does not negate this theory. In addition, our results show that zip regulates rotation, as do the tissue polarity genes, suggesting that these genes at least act in parallel pathways. It is important to note, however, that these studies did not explore interactions with fmi, pk, or stbm. Surprisingly, null alleles of frizzledKD4A (fzKD4A) and dsh3 (the dsh3 allele eliminates both canononical and non-canonical Wnt signaling) enhance this phenotype (Fig. 7 C, D, Table 1). Since both the canonical and non-canonical Wnt signaling pathways share fz and dsh, our results raised the intriguing possibility that zip might act through canonical Wnt signaling to regulate ommatidial orientation. Notably, to date, no connection has been made between canonical Wnt signaling and ommatidial orientation. Indeed, armadillo8, (arm8) an allele of the Drosophila β-catenin homolog that specifically blocks wingless signaling while leaving other functions of the gene intact, dominantly enhances the sev>zipDN phenotype, as do fzKD4A and dsh3 (Fig. 7E, Table 1). Zip therefore appears to act through canonical Wnt signaling, perhaps in cooperation with Drok.
Pupal cell death contributes to ommatidial alignment
There is a discrepancy in the standard deviation for ommatidial rotation at the end of larval development (+/-9, row 15) relative to the adult (+/-4). While ommatidial rotation is the initial and major player in polarizing the retinal epithelium -- it turns ommatidial precursors 88° +/-9 - the ∼5° adjustment to standard deviation indicates rotation is clearly not the sole factor that contributes to the precise alignment of the adult ommatidial lattice. Rather, a second, post-rotation mechanism apparently imposes order on a system that is slightly disorganized.
There are several major patterning events that occur between the end of larval life and adult life that could adjust the lattice: additional cells are recruited into the ommatidium from the pool of undifferentiated interommatidial cells; the pigment cell lattice becomes organized as excess, undifferentiated cells are removed by apoptosis; and the four major cell types that comprise each ommatidium differentiate and produce their specialized features (pigment granules, rhabdomeres, lens material, etc.). Of these events, sorting of the interommatidial lattice seems best suited to influence ommatidial orientation. While cell death is likely the major contributor to this sorting event, recruitment of the primary pigment cells could play a minor role since this process effectively removes (and organizes) two cells per ommatidium from the interommatidial lattice (Wolff and Ready, 1991a). In fact, by 30 hours of pupal life (20°), at which point the primary pigment cells have been recruited and have fully enwrapped the cone cells to establish at least one of their two contacts, the cone cells are clearly more organized, reflecting an increased state of order in the underlying photoreceptors (see Fig. 20 in (Wolff and Ready, 1993)).
To test for a role for cell death in ommatidial alignment, we examined eyes from flies that express the baculovirus anti-apoptotic protein, P35 (Hay et al., 1994), to see if P35 eyes undergo a similar correction of the standard deviation as described for wild type (+/-9 to +/-4, discussed above). P35 expression in the eye blocks cell death, resulting in an eye with excess secondary and tertiary pigment cells (Hay et al., 1994). Adult P35 eyes exhibit orientation defects, including those that fall below 90° as well as those that surpass 90° (91° +/- 12). The standard deviation at row 15 in P35 eye discs is +/-13 (Fig. 7F, Fig. 3). Although the standard deviation in P35 eye discs in row 15 is greater than the wild-type equivalent, the failure to improve (i.e. decrease) the standard deviation between larval and adult life in P35 eyes suggests the removal of extra cells contributes to the resolution of the cellular network into its widely recognized honeycomb lattice.
This transgene also has a modest effect on rotation in that initiation is delayed by one row. The effect may simply be a non-specific consequence of overexpressing a protein in the eye, as sev>zip exhibits a similar phenotype. However, it is difficult to imagine that this slight delay could disrupt ommatidial orientation to the extent seen in the adult eye.
Cell death genes modify the zip ommatidial orientation phenotype
The standard deviation for ommatidial orientation does not decrease between larval and adult eyes in P35 eyes, a phenomenon that we propose is, at least in part, controlled by the selective removal of cells via apoptosis. Similarly, the standard deviation for sev>zipDN does not change between these two time points (Fig. 3, Table 1). Furthermore, we show that zip interacts with members of the canonical Wnt signaling pathway, a pathway known to regulate cell death (Ahmed et al., 1998; Cox et al., 2000; Freeman and Bienz, 2001). These observations raised the possibility that zip may play a role in regulating cell death to adjust ommatidial alignment independently of its effect on rotation. We therefore tested the deficiency line (Df (H99) kind gift of R. Cagan) which uncovers three genes that promote cell death: grim, reaper (rpr) and head involuted (hid) (Foley and Cooley, 1998), as well as a mutation in hid, for their ability to modify the sev>zipDN phenotype. Indeed, both the deficiency and hid05014 suppress the sev>zipDN phenotype (Fig. 7G, H; Table 1). One hypothesis that is supported by these data is that zip plays a second, independent and post-rotation role in ommatidial rotation by contributing to patterning the pigment cell lattice via cell death. As a first step in exploring this possible role, we characterized the mid-pupal eye phenotype of loss-of-function alleles of zip.
The network of bristles and secondary and tertiary pigment cells is disrupted in partial loss of zip function mutants. The complement and arrangement of cone cells and primary pigment cells, on the other hand, is normal. In wild-type eyes, bristles and tertiary pigment cells occupy alternate vertices, and each of these cells contacts three primary pigment cells (Fig. 8A). The secondary pigment cells lie along each of the six faces of the hexagon and contact a bristle cell at one end and a tertiary pigment cell at the other end. When zip function is decreased in zipEbr/zip 02957 (Fig. 8B) or using the zipDN transgene driven by sev (Fig. 8C), multiple defects are seen, including a failure to sort cells at vertices (Fig. 8B, C, single arrowheads), cells that expand to occupy more than one niche (Fig. 8B, arrow: this bristle is situated roughly at the vertex but also stretches out to occupy a position normally filled by a secondary pigment cell), extra cells (Fig. 8B, double arrow), and missing cells (Fig. 8C, arrow). Whether the disruptions in the pigment cell lattice are a direct consequence of a role for zip in regulating cell death or an indirect consequence of earlier rotation defects, is unclear (see Discussion).
Figure 8. The interommatidial lattice is disrupted when Zip function is compromised.
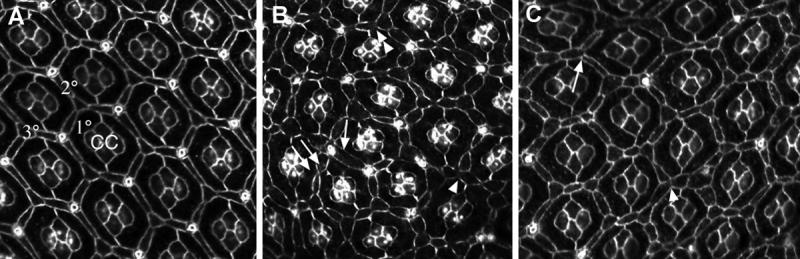
(A) Wild-type mid-pupal eye stained with α-Arm. The interommatidial lattice in wild-type eyes is virtually error-free. 1°, 2°, 3°: primary, secondary, and tertiary pigment cells, respectively; cc: cone cells. Bristles cells appear as white circles in these preparations. zipEbr/zip02957 (B) and sev>zipDN (C) mid-pupal eyes stained with α -Arm. Multiple patterning defects arise when Zip function is decreased. See text for details.
Finally, primary pigment cell boundaries often have scalloped edges and α-Arm staining is frequently discontinuous, giving the impression that the adherens junctions are disintegrating; in many cases, it looks as if cells have pulled away from each other (double arrowhead, Fig. 8B). While the scalloped morphology and discontinous α-Arm pattern both occur in wild type, they are only seen in significantly younger eyes.
Discussion
The mechanisms that drive rotation and how their output translates into the sophisticated morphogenetic movements that give rise to polarized epithelia are poorly understood. The synchronized coordination of numerous systems likely comprises the driving force, or “motor,” for rotation. The force could be a consequence of coordinated changes in cell adhesion between appropriate subsets of cells. Indeed, such a model has been proposed by Mlodzik, in which they demonstrate roles for DE-Cadherin and DN-Cadherin in ommatidial rotation (Mirkovic and Mlodzik, 2006). Alternatively, the force to move the cells past one another could be actino-myosin-based, as occurs in CE (Bertet et al., 2004). The output of the force may in turn, drive the remodeling of adherens junctions to move cells past one another, as theorized for CE (Bertet et al., 2004). The force could be more of a pulling force, for example, by filopodia, in which microtubules slide past one another to pull one set of cells, the photoreceptors (and later cone cells), past a second set, the undifferentiated cells. In conjunction with force generation, it is expected that intercellular contacts between the two subsets of cells would be remodeled to enable ommatidial precursors to rotate within the retinal primordium without disrupting the integrity and overall shape of the epithelium. All systems and molecules involved in junctional remodeling should therefore also be considered components of the motor for rotation.
Several observations reported here indicate that Zip contributes to the regulation of rotation. Misexpression of both wild-type zip and a dominant negative form of zip, which effectively knocks down Zip function, as well as loss-of-function zip, interfere with normal rotation. The mechanical forces generated by Zip regulate dorsal closure, cell locomotion and morphogenetic movements (Young et al., 1993) and they modulate the changes in cell-cell junctions that drive the cellular movements underlying CE (Bertet et al., 2004). It is unclear what role Zip is fulfilling within the apical domain of rotating ommatidia, but several possibilities, based on the identified functions for non-muscle myosin in diverse developmental events, are proposed. Non-muscle myosin transports molecules to asymmetrically localize them within cells (Barros et al., 2003; Huang et al., 2003; Jin et al., 2001). Zip could function in this capacity in the Drosophila eye to direct relevant molecules, for example tissue polarity or adhesion molecules, to appropriate regions within the cell. In other events, Zip produces the force for apical constriction (Dietz et al., 2006), so conceivably, a directed cell shape change in a group of cells could produce the rotational force. However, this model is unlikely given that the changes in cell shape during rotation are minimal and intuitively seem insufficient to account for the vast distance the ommatidia move. Finally, Zip may act in the same capacity as it does in CE. In CE, Zip is proposed to drive the remodeling of adherens junctions by generating local forces at cell boundaries, thereby pulling opposing cells together (Zallen and Wieschaus, 2004). While there are differences between CE and rotation, polarized morphogenesis underlies both events and parallel mechanisms could regulate both processes. In rotation, Zip may create local forces in rotating cells or in rotating vs. non-rotating cells as a means of remodeling the adherens junctions, ultimately pulling dynamic cells past their stationary neighbors.
We favor a model in which rotation represents a modified form of CE given 1) the similarities in the polarized cell movements between CE and rotation, and 2) there are overlapping genes required in both processes. In this model, we propose that Zip generates a force within moving cells to move them past stationary cells during ommatidial rotation. This model is modified to account for the fact that whereas all cells move in CE, only a subset of cells moves in rotation. Furthermore, to accommodate eye development, the force would have to be unidirectional and that direction of force would be opposite in the dorsal and ventral halves of the eye, with the direction likely being controlled by the tissue polarity complex. We propose that Zip, likely through its interactions with the dense accumulations of actin that appear at the onset of rotation, is essential for movement. The observations reported here are consistent with this model in that actin and Zip materialize coincident with rotation and they do so only in moving cells.
zip mutants exhibit a significant patterning defect in mid-pupal eyes, but the precise role for zip following the completion of rotation is unclear. Cell fates in the eye are determined based on contacts with adjacent cells. During pupal eye development, undifferentiated interommatidial cells are directed to differentiate or die, depending on the set of contacts they establish with their neighbors. By row 15 of sev>zipDN eye discs, many ommatidial precursors are significantly off-axis in their alignment, a phenotype that does not get corrected during later stages of development (standard deviation: +/- 20). The ommatidial core on which future cell sorting is based is consequently disorganized, precluding the ability to achieve the appropriate contacts to order the interommatidial network of cells. By virtue of its rotation phenotype, zip therefore indirectly affects cell death.
The aberrant patterning and resulting aberrant cell death may also account for the over-rotated ommatidia in zip mutants. If the local environment of an ommatidium is relatively abnormal, then the shifting that occurs as cell death removes cells could exacerbate the phenotype, causing some ommatidia to move past 90°.
In addition to this predicted, indirect effect on cell death, the genetic data presented here suggest that zip could also act with canonical Wnt signaling to regulate ommatidial orientation: members of the canonical Wnt signaling pathway (fzKD4A, arm8 and dsh3), which promotes apoptosis in the eye during pupal life (Ahmed et al., 1998; Ahmed et al., 2002; Brunner et al., 1999; Freeman and Bienz, 2001), enhance the zip phenotype. Future work should provide an understanding of the link and nature of the interactions between zip, canonical signaling and cell death genes.
Acknowledgements
We express our gratitude to K. Choi, E. Verheyen, D. Kiehart, R. Ward, A. Brand, R. Cagan and the Bloomington Stock Center for fly stocks, D. Kiehart for generously providing antibodies, and S. Takeda and M. Das Thakur for research assistance. We also thank D. Ready for thoughtful and provocative discussions and members of the Wolff lab for their invaluable input. This investigation was supported by National Institutes of Health, Institutional National Research Service Award 5-T32-EY13360-05 to R.W.F. and NIH grant R01 EY13136 to T.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed Y, et al. Regulation of armadillo by a Drosophila APC inhibits neuronal apoptosis during retinal development. Cell. 1998;93:1171–82. doi: 10.1016/s0092-8674(00)81461-0. [DOI] [PubMed] [Google Scholar]
- Ahmed Y, et al. Drosophila Apc1 and Apc2 regulate Wingless transduction throughout development. Development. 2002;129:1751–62. doi: 10.1242/dev.129.7.1751. [DOI] [PubMed] [Google Scholar]
- Baker NE, et al. Evolution of proneural atonal expression during distinct regulatory phases in the developing Drosophila eye. Curr Biol. 1996;6:1290–301. doi: 10.1016/s0960-9822(02)70715-x. [DOI] [PubMed] [Google Scholar]
- Barros CS, et al. Drosophila nonmuscle myosin II promotes the asymmetric segregation of cell fate determinants by cortical exclusion rather than active transport. Dev Cell. 2003;5:829–40. doi: 10.1016/s1534-5807(03)00359-9. [DOI] [PubMed] [Google Scholar]
- Baumann O. Spatial pattern of nonmuscle myosin-II distribution during the development of the Drosophila compound eye and implications for retinal morphogenesis. Dev Biol. 2004;269:519–33. doi: 10.1016/j.ydbio.2004.01.047. [DOI] [PubMed] [Google Scholar]
- Bertet C, et al. Myosin-dependent junction remodelling controls planar cell intercalation and axis elongation. Nature. 2004;429:667–71. doi: 10.1038/nature02590. [DOI] [PubMed] [Google Scholar]
- Brown KE, Freeman M. Egfr signalling defines a protective function for ommatidial orientation in the Drosophila eye. Development. 2003;130:5401–12. doi: 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- Brunner E, et al. The dominant mutation Glazed is a gain-of-function allele of wingless that, similar to loss of APC, interferes with normal eye development. Dev Biol. 1999;206:178–88. doi: 10.1006/dbio.1998.9136. [DOI] [PubMed] [Google Scholar]
- Burns CG, et al. Expression of light meromyosin in Dictyostelium blocks normal myosin II function. J Cell Biol. 1995;130:605–12. doi: 10.1083/jcb.130.3.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos-Ortega J. A. a. A. H. Cell Clones and pattern formation: On the lineage of photoreceptor cells in the compound eye of Drosophila. Wilhelm Roux’s Archives of Developmental Biology. 1977;181:227–245. doi: 10.1007/BF00848423. [DOI] [PubMed] [Google Scholar]
- Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78:125–36. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- Chou YH, Chien CT. Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev Cell. 2002;3:839–50. doi: 10.1016/s1534-5807(02)00362-3. [DOI] [PubMed] [Google Scholar]
- Cox RT, et al. A screen for mutations that suppress the phenotype of Drosophila armadillo, the beta-catenin homolog. Genetics. 2000;155:1725–40. doi: 10.1093/genetics/155.4.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz ML, et al. Differential actin-dependent localization modulates the evolutionarily conserved activity of Shroom family proteins. J Biol Chem. 2006;281:20542–54. doi: 10.1074/jbc.M512463200. [DOI] [PubMed] [Google Scholar]
- Djiane A, et al. Role of frizzled 7 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development. 2000;127:3091–100. doi: 10.1242/dev.127.14.3091. [DOI] [PubMed] [Google Scholar]
- Dokucu ME, et al. Atonal, rough and the resolution of proneural clusters in the developing Drosophila retina. Development. 1996;122:4139–47. doi: 10.1242/dev.122.12.4139. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Kiehart DP. Drosophila nonmuscle myosin II has multiple essential roles in imaginal disc and egg chamber morphogenesis. Development. 1996;122:1499–511. doi: 10.1242/dev.122.5.1499. [DOI] [PubMed] [Google Scholar]
- Fanto M, et al. Nuclear signaling by Rac and Rho GTPases is required in the establishment of epithelial planar polarity in the Drosophila eye. Curr Biol. 2000;10:979–88. doi: 10.1016/s0960-9822(00)00645-x. [DOI] [PubMed] [Google Scholar]
- Field CM, Alberts BM. Anillin, a contractile ring protein that cycles from the nucleus to the cell cortex. J Cell Biol. 1995;131:165–78. doi: 10.1083/jcb.131.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley K, Cooley L. Apoptosis in late stage Drosophila nurse cells does not require genes within the H99 deficiency. Development. 1998;125:1075–82. doi: 10.1242/dev.125.6.1075. [DOI] [PubMed] [Google Scholar]
- Formstone CJ, Mason I. Expression of the Celsr/flamingo homologue, c-fmi1, in the early avian embryo indicates a conserved role in neural tube closure and additional roles in asymmetry and somitogenesis. Dev Dyn. 2005;232:408–13. doi: 10.1002/dvdy.20228. [DOI] [PubMed] [Google Scholar]
- Franke JD, et al. Nonmuscle myosin II generates forces that transmit tension and drive contraction in multiple tissues during dorsal closure. Curr Biol. 2005;15:2208–21. doi: 10.1016/j.cub.2005.11.064. [DOI] [PubMed] [Google Scholar]
- Freeman M, Bienz M. EGF receptor/Rolled MAP kinase signalling protects cells against activated Armadillo in the Drosophila eye. EMBO Rep. 2001;2:157–62. doi: 10.1093/embo-reports/kve019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman M, et al. The argos gene encodes a diffusible factor that regulates cell fate decisions in the Drosophila eye. Cell. 1992;69:963–75. doi: 10.1016/0092-8674(92)90615-j. [DOI] [PubMed] [Google Scholar]
- Fulga TA, Rorth P. Invasive cell migration is initiated by guided growth of long cellular extensions. Nat Cell Biol. 2002;4:715–9. doi: 10.1038/ncb848. [DOI] [PubMed] [Google Scholar]
- Gaengel K, Mlodzik M. Egfr signaling regulates ommatidial rotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development. 2003;130:5413–23. doi: 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- Hay BA, et al. Expression of baculovirus P35 prevents cell death in Drosophila. Development. 1994;120:2121–9. doi: 10.1242/dev.120.8.2121. [DOI] [PubMed] [Google Scholar]
- Huang H, et al. Nonmuscle myosin promotes cytoplasmic localization of PBX. Mol Cell Biol. 2003;23:3636–45. doi: 10.1128/MCB.23.10.3636-3645.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa R, et al. Polarized actin bundles formed by human fascin-1: their sliding and disassembly on myosin II and myosin V in vitro. J Neurochem. 2003;87:676–85. doi: 10.1046/j.1471-4159.2003.02058.x. [DOI] [PubMed] [Google Scholar]
- Jin Y, et al. Myosin ii light chain phosphorylation regulates membrane localization and apoptotic signaling of tumor necrosis factor receptor-1. J Biol Chem. 2001;276:30342–9. doi: 10.1074/jbc.M102404200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltschmidt JA, et al. Planar polarity and actin dynamics in the epidermis of Drosophila. Nat Cell Biol. 2002;4:937–44. doi: 10.1038/ncb882. [DOI] [PubMed] [Google Scholar]
- Kiehart DP, Feghali R. Cytoplasmic myosin from Drosophila melanogaster. J Cell Biol. 1986;103:1517–25. doi: 10.1083/jcb.103.4.1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuczmarski ER, et al. Stopped-flow measurement of cytoskeletal contraction: Dictyostelium myosin II is specifically required for contraction of amoeba cytoskeletons. J Cell Biol. 1991;114:1191–9. doi: 10.1083/jcb.114.6.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EC, et al. The scabrous gene encodes a secreted glycoprotein dimer and regulates proneural development in Drosophila eyes. Mol Cell Biol. 1996;16:1179–88. doi: 10.1128/mcb.16.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locascio A, Nieto MA. Cell movements during vertebrate development: integrated tissue behaviour versus individual cell migration. Curr Opin Genet Dev. 2001;11:464–9. doi: 10.1016/s0959-437x(00)00218-5. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, et al. Drosophila nemo is an essential gene involved in the regulation of programmed cell death. Mech Dev. 2002;119:9–20. doi: 10.1016/s0925-4773(02)00289-7. [DOI] [PubMed] [Google Scholar]
- Mirkovic I, Mlodzik M. Cooperative activities of drosophila DE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development. 2006;133:3283–93. doi: 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- Mizuno T, et al. Drosophila myosin phosphatase and its role in dorsal closure. Development. 2002;129:1215–23. doi: 10.1242/dev.129.5.1215. [DOI] [PubMed] [Google Scholar]
- Mlodzik M. Planar cell polarization: do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation? Trends Genet. 2002;18:564–71. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- Montell DJ. Border-cell migration: the race is on. Nat Rev Mol Cell Biol. 2003;4:13–24. doi: 10.1038/nrm1006. [DOI] [PubMed] [Google Scholar]
- Morel V, Arias AM. Armadillo/beta-catenin-dependent Wnt signalling is required for the polarisation of epidermal cells during dorsal closure in Drosophila. Development. 2004;131:3273–83. doi: 10.1242/dev.01217. [DOI] [PubMed] [Google Scholar]
- Ready DF, et al. Development of the Drosophila retina, a neurocrystalline lattice. Dev Biol. 1976;53:217–40. doi: 10.1016/0012-1606(76)90225-6. [DOI] [PubMed] [Google Scholar]
- Strutt H, Strutt D. EGF signaling and ommatidial rotation in the Drosophila eye. Curr Biol. 2003;13:1451–7. doi: 10.1016/s0960-9822(03)00545-1. [DOI] [PubMed] [Google Scholar]
- Tada M, et al. Non-canonical Wnt signalling and regulation of gastrulation movements. Semin Cell Dev Biol. 2002;13:251–60. doi: 10.1016/s1084-9521(02)00052-6. [DOI] [PubMed] [Google Scholar]
- Tomlinson A. The cellular dynamics of pattern formation in the eye of Drosophila. J Embryol Exp Morphol. 1985;89:313–31. [PubMed] [Google Scholar]
- Tomlinson A, et al. Localization of the sevenless protein, a putative receptor for positional information, in the eye imaginal disc of Drosophila. Cell. 1987;51:143–50. doi: 10.1016/0092-8674(87)90019-5. [DOI] [PubMed] [Google Scholar]
- Verdier V, et al. Rho-kinase regulates tissue morphogenesis via non-muscle myosin and LIM-kinase during Drosophila development. BMC Dev Biol. 2006;6:38. doi: 10.1186/1471-213X-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward RE, et al. Genetic modifier screens in Drosophila demonstrate a role for Rho1 signaling in ecdysone-triggered imaginal disc morphogenesis. Genetics. 2003;165:1397–415. doi: 10.1093/genetics/165.3.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winklbauer R, et al. Frizzled-7 signalling controls tissue separation during Xenopus gastrulation. Nature. 2001;413:856–60. doi: 10.1038/35101621. [DOI] [PubMed] [Google Scholar]
- Winter CG, et al. Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton. Cell. 2001;105:81–91. doi: 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- Wolff T. Histological Techniques for the Drosophila Eye. In: Sullivan William, Ashburner Michael, Scott Hawley R., editors. Drosophila Protocols. Cold Spring Harbor Press; Cold Spring Harbor, NY: 2000. pp. 201–244. [Google Scholar]
- Wolff T, Ready DF. Cell death in normal and rough eye mutants of Drosophila. Development. 1991a;113:825–39. doi: 10.1242/dev.113.3.825. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development. 1991b;113:841–50. doi: 10.1242/dev.113.3.841. [DOI] [PubMed] [Google Scholar]
- Wolff T, Ready DF. Pattern formationin the Drosophila retina. In: Bate Michael, Martinez-Arias Alfonso., editors. The Development of Drosophila melanogaster. Cold Spring Harbor Press; Cold Spring Harbor, NY: 1993. pp. 1277–1325. [Google Scholar]
- Young PE, et al. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 1993;7:29–41. doi: 10.1101/gad.7.1.29. [DOI] [PubMed] [Google Scholar]
- Zallen JA, Wieschaus E. Patterned gene expression directs bipolar planar polarity in Drosophila. Dev Cell. 2004;6:343–55. doi: 10.1016/s1534-5807(04)00060-7. [DOI] [PubMed] [Google Scholar]