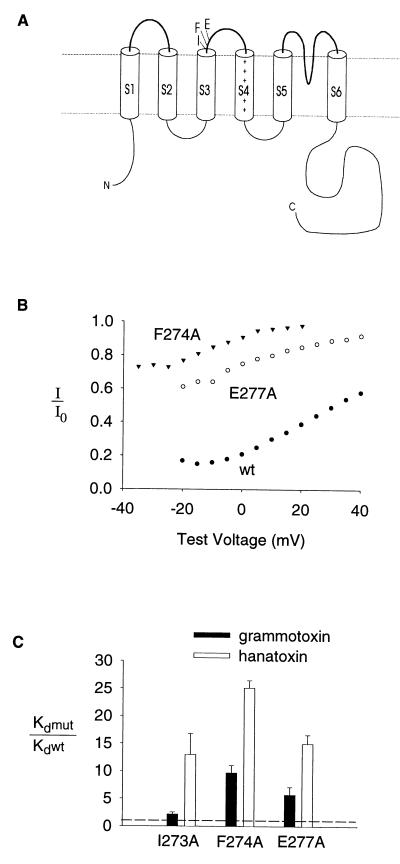
Figure 4.
Mutation of residues in the S3–S4 linker alters the binding affinity of grammotoxin to the drk1 K+ channel. (A) Membrane folding model of a single K+ channel α subunit showing the location of three residues at the C-terminal end of S3 that influence hanatoxin-binding affinity (14). (B) Fraction of uninhibited tail current elicited by various strength depolarizations at 10 μM grammotoxin for F274A, E277A and the wild-type channel. Tail current amplitude in the presence of 10 μM grammotoxin (I) and tail current in control (Io). All tail currents were elicited by repolarization to −50 mV. Test depolarizations were 250 msec in duration from a holding voltage of −80 mV. (C) Equilibrium dissociation constants (Kd) for grammotoxin binding to mutant drk1 K+ channels shown as a fraction of the Kd for grammotoxin binding to the wild-type drk1 K+ channel. Kd values for grammotoxin (mean ± SEM; n = 3–5) were calculated from the fraction of uninhibited current at negative voltages as previously described for hanatoxin (13) and shown in Fig. 2 and legend. Data for hanatoxin from ref. 14 are shown for comparison.