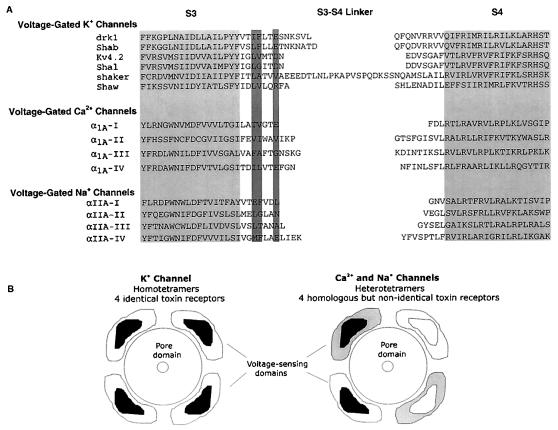
Figure 5.
(A) Alignment of various voltage-gated ion channels in a region spanning from S3 through S4. Light shading indicates proposed transmembrane segments. Dark shading highlights the position of residues that when mutated in the drk1 K+ channel alters hanatoxin- and grammotoxin-binding affinity (14, Fig. 4). Amino acid sequence for α1A is from ref. 23, and the sequence for αIIA is from ref. 42. (B) Diagram illustrating voltage-sensing domains (with toxin receptors contained within) and some differences that likely exist between voltage-gated K+, Ca2+, and Na+ channels. Voltage-sensing domains are drawn as modules that surround the pore domain. K+ channels studied experimentally are tetramers of four identical subunits. Each subunit contains a voltage-sensing domain to which a toxin binds. The toxin receptor is located at least 10–15 Å from the central pore axis. Ca2+ and Na+ channels are heterotetramers that contain four homologous but different pseudosubunits, each containing a voltage-sensing domain that likely binds gating modifier toxins with different energetics. Toxin receptors are arbitrarily drawn as contained within each subunit; receptors could be at the interface between subunits.