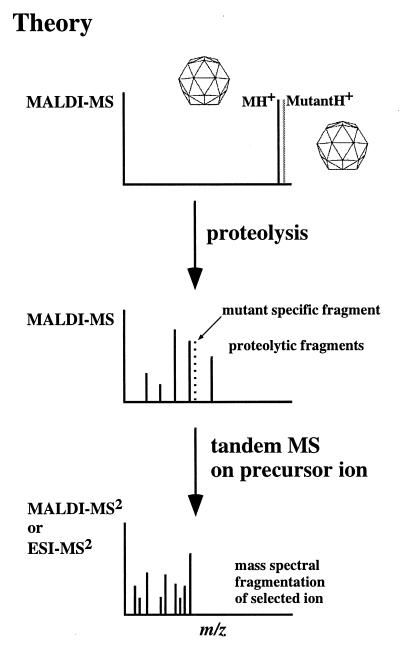
Figure 1.
Schematic representation of the method to identify virus mutants by mass spectrometry. (Top) The observation of differences between wild-type and mutant viruses indicates amino acid changes (i.e., mutations). (Middle) Proteolysis of the viral variants generates a set of digestion fragments (one or more of which may contain the mutated amino acid), which appear at different m/z values as a consequence of an amino acid change(s). This observation provides valuable information as to the location of the mutation, augmenting its identification either by further mass spectral analysis (Bottom spectra) or nucleotide sequencing. (Bottom) Tandem mass spectrometric experiments (on the fragment containing the mutation) generate sequence information providing definitive identification of the mutated amino acid.