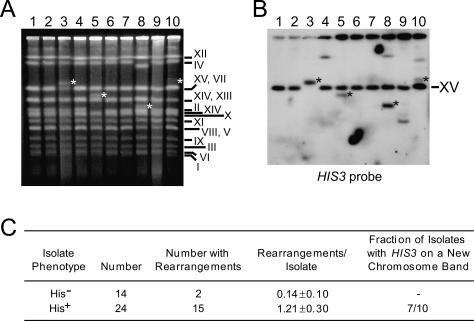
Figure 2.
Selection for ADH1HIS3 retrosequence formation is correlated with the appearance of chromosome rearrangements. (A) Lanes 1–5 and 6–10 each show intact chromosomes from five clones obtained from an independent His− telomerase-negative survivor separated using CHEF gel electrophoresis. Two clones in each set were obtained without selection for retrosequence formation (lanes 1,2,6,7) and therefore remained His−, while three clones in each set were His+ due to selection for retrosequences (lanes 3–5,8–10). Asterisks indicate new chromosome bands that also harbored HIS3 sequences (see below). The yeast chromosome or pair of chromosomes corresponding to each band is indicated to the right of the gel image. (B) A Southern blot of the gel shown in A was probed for HIS3 sequences. Both ADH1his3AI and a partial deletion allele of HIS3, his3Δ1, are present on chromosome XV, so the band corresponding to chromosome XV was detected in all lanes. (Lanes 3–5,8–10) At least one additional band was detected in each sample from His+ clones, with the exception of the sample in lane 3. The lack of an additional band in lane 3 resulted from the incorporation of ADH1HIS3 into chromosome XV. Asterisks indicate the same new chromosome bands indicated in A. (C) Chromosomal rearrangement frequencies in isolates obtained from a total of three independent His− telomerase-negative survivors. New bands or missing bands on CHEF gels were scored as rearrangements. The mean (±standard error) number of rearrangements per isolate is shown, and the difference in the means is statistically significant (P = 0.001).