Abstract
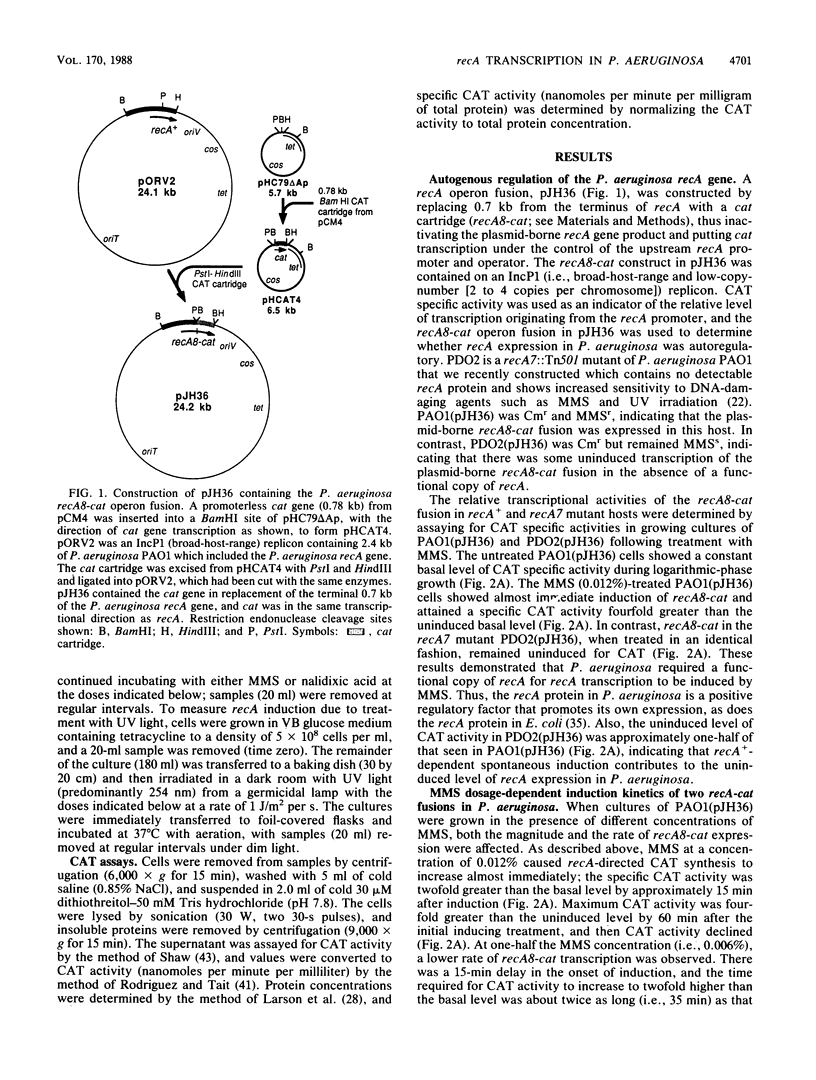
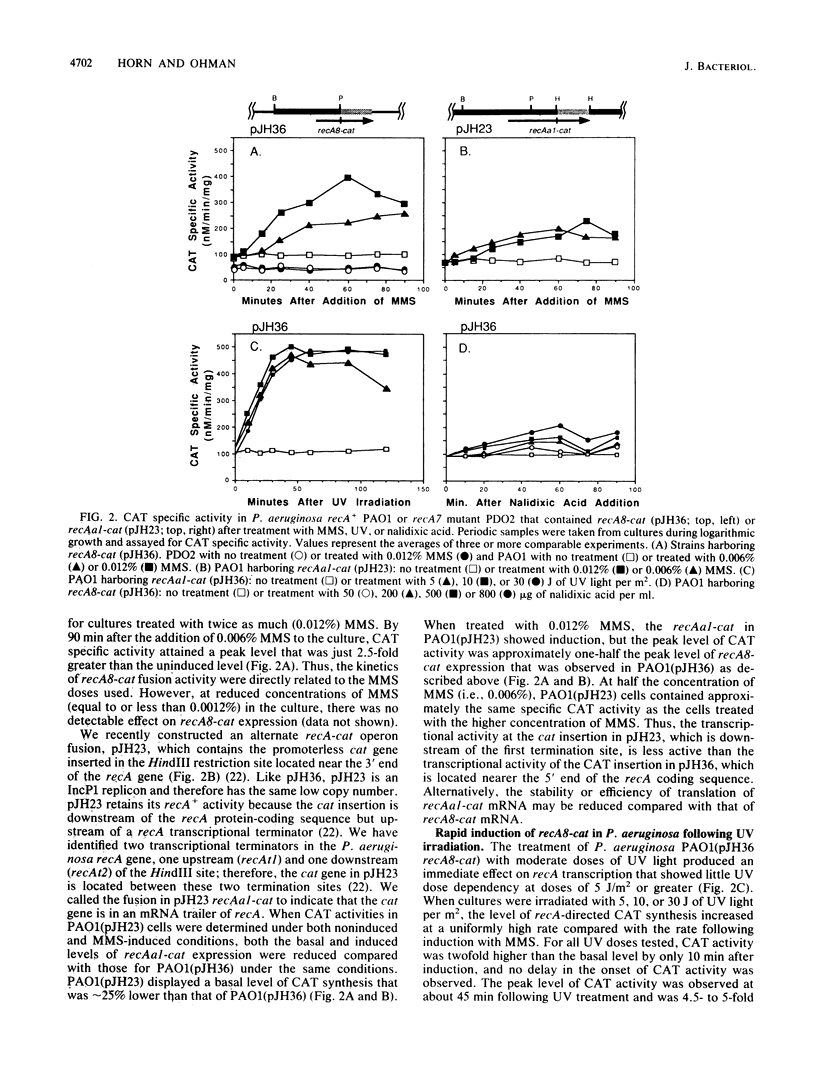
A promoterless chloramphenicol acetyltransferase gene (cat) was used to construct recA-cat operon fusions to quantitatively examine the transcriptional regulation of the Pseudomonas aeruginosa recA gene in P. aeruginosa PAO. Wild-type P. aeruginosa containing the recA8-cat fusion was treated with methyl methanesulfonate (MMS) and showed immediate induction of chloramphenicol acetyltransferase (CAT) specific activity, whereas a recA::Tn501 mutant of P. aeruginosa containing recA8-cat showed no induction with MMS. This indicated that a functional copy of recA was required for derepression of recA transcription and that P. aeruginosa recA protein was a positive regulatory factor promoting its own expression. Compared with that in the wild type, the uninduced level of CAT in recA8-cat-containing cells was reduced by approximately one-half in the recA::Tn501 mutant, indicating that recA+-dependent spontaneous induction contributes to the uninduced levels of recA expression in P. aeruginosa. MMS (0.012%) caused recA-directed CAT synthesis to increase almost immediately, with maximum CAT activity, fourfold higher than uninduced levels, attained at 60 min postinduction. The kinetics of recA8-cat fusion activity were shown to be directly related to the MMS doses used. Another fusion called recAa1-cat, where cat was located between the two transcriptional terminators of the P. aeruginosa recA gene, also showed dose-dependent induction by MMS, but the CAT activity from recAa1-cat was only one-half of that obtained with recA8-cat under the same conditions. Treatment of recA+ P. aeruginosa containing recA8-cat with UV irradiation produced an immediate effect on recA8-cat transcription and showed little UV dose dependency at doses of 5 J/m2 or greater. Treatment with 10 J/m2 produced peak levels of recA-directed CAT activity, fivefold higher than background levels, by 60 min postirradiation; CAT activity remained at peak levels during the 120 min of the experiment. In contrast, nalidixic acid had a weak effect on recA8-cat expression in P. aeruginosa, although the response was dose dependent. Nalidixic acid (800 micrograms/ml) produced maximal CAT activity that was only twofold higher than background levels.
Full text
PDF




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. Mechanism of action of the lexA gene product. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4204–4208. doi: 10.1073/pnas.78.7.4204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaregola S., D'Ari R., Huisman O. Quantitative evaluation of recA gene expression in Escherichia coli. Mol Gen Genet. 1982;185(3):430–439. doi: 10.1007/BF00334135. [DOI] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Close T. J., Rodriguez R. L. Construction and characterization of the chloramphenicol-resistance gene cartridge: a new approach to the transcriptional mapping of extrachromosomal elements. Gene. 1982 Dec;20(2):305–316. doi: 10.1016/0378-1119(82)90048-8. [DOI] [PubMed] [Google Scholar]
- Cohen S. N., Chang A. C., Hsu L. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc Natl Acad Sci U S A. 1972 Aug;69(8):2110–2114. doi: 10.1073/pnas.69.8.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig N. L., Roberts J. W. E. coli recA protein-directed cleavage of phage lambda repressor requires polynucleotide. Nature. 1980 Jan 3;283(5742):26–30. doi: 10.1038/283026a0. [DOI] [PubMed] [Google Scholar]
- Dressler D., Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eitner G., Adler B., Lanzov V. A., Hofemeister J. Interspecies recA protein substitution in Escherichia coli and Proteus mirabilis. Mol Gen Genet. 1982;185(3):481–486. doi: 10.1007/BF00334144. [DOI] [PubMed] [Google Scholar]
- Figurski D. H., Helinski D. R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci U S A. 1979 Apr;76(4):1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J. L., Ohman D. E. Cloning of genes from mucoid Pseudomonas aeruginosa which control spontaneous conversion to the alginate production phenotype. J Bacteriol. 1988 Apr;170(4):1452–1460. doi: 10.1128/jb.170.4.1452-1460.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman A. M., Long S. R., Brown S. E., Buikema W. J., Ausubel F. M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982 Jun;18(3):289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- Goldberg J. B., Ohman D. E. Cloning and expression in Pseudomonas aeruginosa of a gene involved in the production of alginate. J Bacteriol. 1984 Jun;158(3):1115–1121. doi: 10.1128/jb.158.3.1115-1121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanawalt P. C., Cooper P. K., Ganesan A. K., Smith C. A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- Hohn B., Collins J. A small cosmid for efficient cloning of large DNA fragments. Gene. 1980 Nov;11(3-4):291–298. doi: 10.1016/0378-1119(80)90069-4. [DOI] [PubMed] [Google Scholar]
- Holloway B. W., Krishnapillai V., Morgan A. F. Chromosomal genetics of Pseudomonas. Microbiol Rev. 1979 Mar;43(1):73–102. doi: 10.1128/mr.43.1.73-102.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horii T., Ogawa T., Nakatani T., Hase T., Matsubara H., Ogawa H. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell. 1981 Dec;27(3 Pt 2):515–522. doi: 10.1016/0092-8674(81)90393-7. [DOI] [PubMed] [Google Scholar]
- Horn J. M., Ohman D. E. Transcriptional and translational analyses of recA mutant alleles in Pseudomonas aeruginosa. J Bacteriol. 1988 Apr;170(4):1637–1650. doi: 10.1128/jb.170.4.1637-1650.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keener S. L., McNamee K. P., McEntee K. Cloning and characterization of recA genes froM Proteus vulgaris, Erwinia carotovora, Shigella flexneri, and Escherichia coli B/r. J Bacteriol. 1984 Oct;160(1):153–160. doi: 10.1128/jb.160.1.153-160.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C. J., Brent R., Ptashne M., Walker G. C. Regulation of damage-inducible genes in Escherichia coli. J Mol Biol. 1982 Sep 25;160(3):445–457. doi: 10.1016/0022-2836(82)90307-2. [DOI] [PubMed] [Google Scholar]
- Kenyon C. J., Walker G. C. DNA-damaging agents stimulate gene expression at specific loci in Escherichia coli. Proc Natl Acad Sci U S A. 1980 May;77(5):2819–2823. doi: 10.1073/pnas.77.5.2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khidhir M. A., Casaregola S., Holland I. B. Mechanism of transient inhibition of DNA synthesis in ultraviolet-irradiated E. coli: inhibition is independent of recA whilst recovery requires RecA protein itself and an additional, inducible SOS function. Mol Gen Genet. 1985;199(1):133–140. doi: 10.1007/BF00327522. [DOI] [PubMed] [Google Scholar]
- Kogoma T., Torrey T. A., Connaughton M. J. Induction of UV-resistant DNA replication in Escherichia coli: induced stable DNA replication as an SOS function. Mol Gen Genet. 1979 Oct 2;176(1):1–9. doi: 10.1007/BF00334288. [DOI] [PubMed] [Google Scholar]
- Larson E., Howlett B., Jagendorf A. Artificial reductant enhancement of the Lowry method for protein determination. Anal Biochem. 1986 Jun;155(2):243–248. doi: 10.1016/0003-2697(86)90432-x. [DOI] [PubMed] [Google Scholar]
- Little J. W., Edmiston S. H., Pacelli L. Z., Mount D. W. Cleavage of the Escherichia coli lexA protein by the recA protease. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3225–3229. doi: 10.1073/pnas.77.6.3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J. W., Mount D. W. The SOS regulatory system of Escherichia coli. Cell. 1982 May;29(1):11–22. doi: 10.1016/0092-8674(82)90085-x. [DOI] [PubMed] [Google Scholar]
- Little J. W. The SOS regulatory system: control of its state by the level of RecA protease. J Mol Biol. 1983 Jul 15;167(4):791–808. doi: 10.1016/s0022-2836(83)80111-9. [DOI] [PubMed] [Google Scholar]
- Markham B. E., Harper J. E., Mount D. W. Physiology of the SOS response: kinetics of lexA and recA transcriptional activity following induction. Mol Gen Genet. 1985;198(2):207–212. doi: 10.1007/BF00382997. [DOI] [PubMed] [Google Scholar]
- Markham B. E., Harper J. E., Mount D. W., Sancar G. B., Sancar A., Rupp W. D., Kenyon C. J., Walker G. C. Analysis of mRNA synthesis following induction of the Escherichia coli SOS system. J Mol Biol. 1984 Sep 15;178(2):237–248. doi: 10.1016/0022-2836(84)90142-6. [DOI] [PubMed] [Google Scholar]
- McPartland A., Green L., Echols H. Control of recA gene RNA in E. coli: regulatory and signal genes. Cell. 1980 Jul;20(3):731–737. doi: 10.1016/0092-8674(80)90319-0. [DOI] [PubMed] [Google Scholar]
- Ohman D. E., West M. A., Flynn J. L., Goldberg J. B. Method for gene replacement in Pseudomonas aeruginosa used in construction of recA mutants: recA-independent instability of alginate production. J Bacteriol. 1985 Jun;162(3):1068–1074. doi: 10.1128/jb.162.3.1068-1074.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. C., Watson R. M., Vinograd J. Mapping of closed circular DNAs by cleavage with restriction endonucleases and calibration by agarose gel electrophoresis. Proc Natl Acad Sci U S A. 1977 Mar;74(3):851–855. doi: 10.1073/pnas.74.3.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E. M., Roberts J. W. Induction of SOS functions: regulation of proteolytic activity of E. coli RecA protein by interaction with DNA and nucleoside triphosphate. Cell. 1981 Jul;25(1):259–267. doi: 10.1016/0092-8674(81)90251-8. [DOI] [PubMed] [Google Scholar]
- Radding C. M. Homologous pairing and strand exchange in genetic recombination. Annu Rev Genet. 1982;16:405–437. doi: 10.1146/annurev.ge.16.120182.002201. [DOI] [PubMed] [Google Scholar]
- Sano Y., Kageyama M. The sequence and function of the recA gene and its protein in Pseudomonas aeruginosa PAO. Mol Gen Genet. 1987 Jul;208(3):412–419. doi: 10.1007/BF00328132. [DOI] [PubMed] [Google Scholar]
- Shaw W. V. Chloramphenicol acetyltransferase from chloramphenicol-resistant bacteria. Methods Enzymol. 1975;43:737–755. doi: 10.1016/0076-6879(75)43141-x. [DOI] [PubMed] [Google Scholar]
- Slilaty S. N., Little J. W. Lysine-156 and serine-119 are required for LexA repressor cleavage: a possible mechanism. Proc Natl Acad Sci U S A. 1987 Jun;84(12):3987–3991. doi: 10.1073/pnas.84.12.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith C. L. Response of recA-dependent operons to different DNA damage signals. J Biol Chem. 1985 Aug 25;260(18):10069–10074. [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Vericat J. A., Guerrero R., Barbé J. Effect of alkylating agents on the expression of inducible genes of Escherichia coli. J Gen Microbiol. 1986 Oct;132(10):2677–2684. doi: 10.1099/00221287-132-10-2677. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Walker G. C. Mutagenesis and inducible responses to deoxyribonucleic acid damage in Escherichia coli. Microbiol Rev. 1984 Mar;48(1):60–93. doi: 10.1128/mr.48.1.60-93.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisemann J. M., Funk C., Weinstock G. M. Measurement of in vivo expression of the recA gene of Escherichia coli by using lacZ gene fusions. J Bacteriol. 1984 Oct;160(1):112–121. doi: 10.1128/jb.160.1.112-121.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West S. C., Little J. W. P. mirabilis RecA protein catalyses cleavage of E. coli LexA protein and the lambda repressor in vitro. Mol Gen Genet. 1984;194(1-2):111–113. doi: 10.1007/BF00383505. [DOI] [PubMed] [Google Scholar]
- de Vos W. M., Venema G. Transformation of Bacillus subtilis competent cells: identification of a protein involved in recombination. Mol Gen Genet. 1982;187(3):439–445. doi: 10.1007/BF00332625. [DOI] [PubMed] [Google Scholar]


