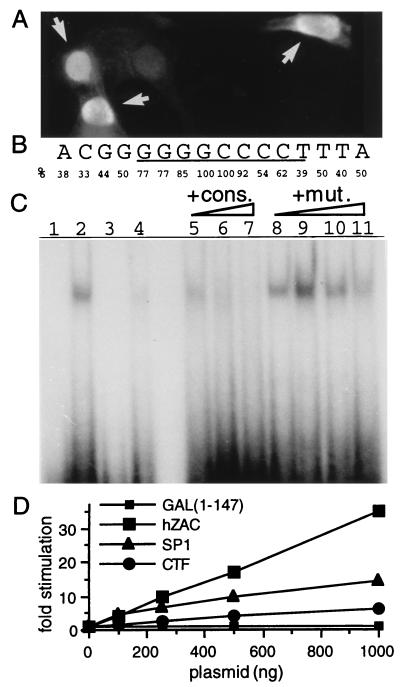
Figure 4.
hZAC intracellular localization, DNA binding, and transactivation. (A) Nuclear localization of hZAC. Immunocytochemistry of SaOs-2 cells transfected with HA-tagged-hZAC (1 μg plasmid) was performed by using an anti-HA antibody and a fluorescein isothiocyanate-conjugated secondary antibody. As cells were transiently transfected, positive transfected cells (indicated with an arrow) as well as negative nontransfected cells were present on the same sample. (B) Consensus nucleotide sequence for hZAC DNA-binding site. This consensus is derived from the alignment of 13 oligonucleotides selected after screening of a random oligonucleotide library. The frequency of the specific base(s) at each position is indicated. (C) Sequence-specific DNA-binding of GST-hZAC-ZF. An electrophoretic mobility-shift assay was performed. GST-hZAC-ZF (100 ng) was allowed to bind to 40,000 cpm of 32P-labeled double-stranded oligonucleotide containing the consensus binding sequence (GGGGGGCCCC; lane 2) or a mutated sequence where the two most conserved bases at the center of the consensus have been mutated (indicated in bold: GGGGGCGCCC; lane 4). Lanes 1 and 3 represent the control electromobilities of both oligonucleotides in the absence of GST-hZAC-ZF. For competition experiments, GST-hZAC-ZF was preincubated with the indicated competitors: consensus (cons.) or mutated consensus (mut.) oligonucleotide and then allowed to bind to the radio-labeled consensus site. Each competitor was added in molar excess of 1-, 10-,100-, and 1,000-fold in lanes 5 and 8, 6 and 9, 7 and 10, and 11, respectively. (D) Transactivation by hZAC. SaOs-2 cells were transfected with plasmids encoding a fusion protein between the GAL4 DNA-binding domain and either hZAC or the transactivation domains of SP1 or CAAT-box binding transcription factor, together with a luciferase reporter gene under the control of a GAL4 sensitive minimal promoter. Luciferase activity for each condition is indicated as a fold stimulation over basal.