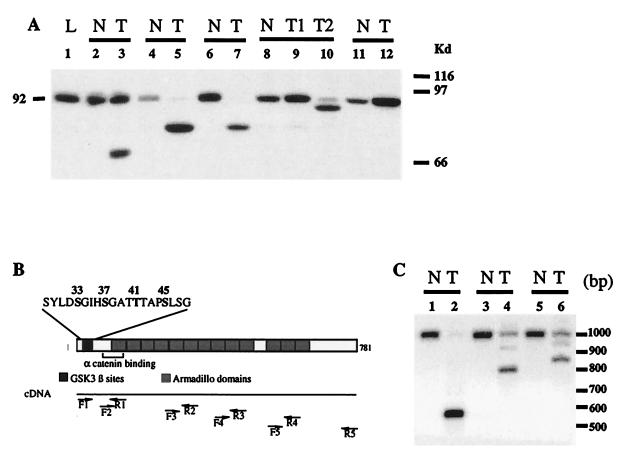
Figure 1.
Analysis of β-catenin in hepatocarcinomas developed in transgenic mice. (A) Immunoblot analysis for β-catenin in L-PK/c-myc transgenic mice. The expected 92-kDa band for β-catenin was detected in normal tissue (N, lanes 2, 4, 6, and 8–11) surrounding the tumor, whereas various truncated proteins were found in tumor nodules (T, lanes 3, 5, 7, 9, and 10). T1 and T2 (lanes 9 and 10) are two independent nodules from the same animal. L, sample from a nontransgenic mouse (lane 1). (B) RT-PCR strategy used to study the β-catenin mRNA. The diagram shows the β-catenin functional domains. The N-terminal domain is involved in the posttranslational stabilization via the GSK-3β phosphorylation site and in binding to α-catenin. The GSK-3β site includes the S33, S37, T41, and S45 phosphorylable residues. The 13 armadillo repeats are shown. Pairs of primers were designed to amplify overlapping PCR products that covered the whole ORF of β-catenin (F1-R1, F2-R2, F3-R3, F4-R4, and F5-R5, see Materials and Methods for the sequence). (C) Southern analysis of RT-PCR showing deleted β-catenin mRNA in L-PK/c-myc transgenic mice. mRNAs obtained from tumor tissue (lanes 2, 4, and 6) together with their nontumor counterparts (lanes 1, 3, and 5) were analyzed by RT-PCR with primers F1-R2, followed by Southern analysis.