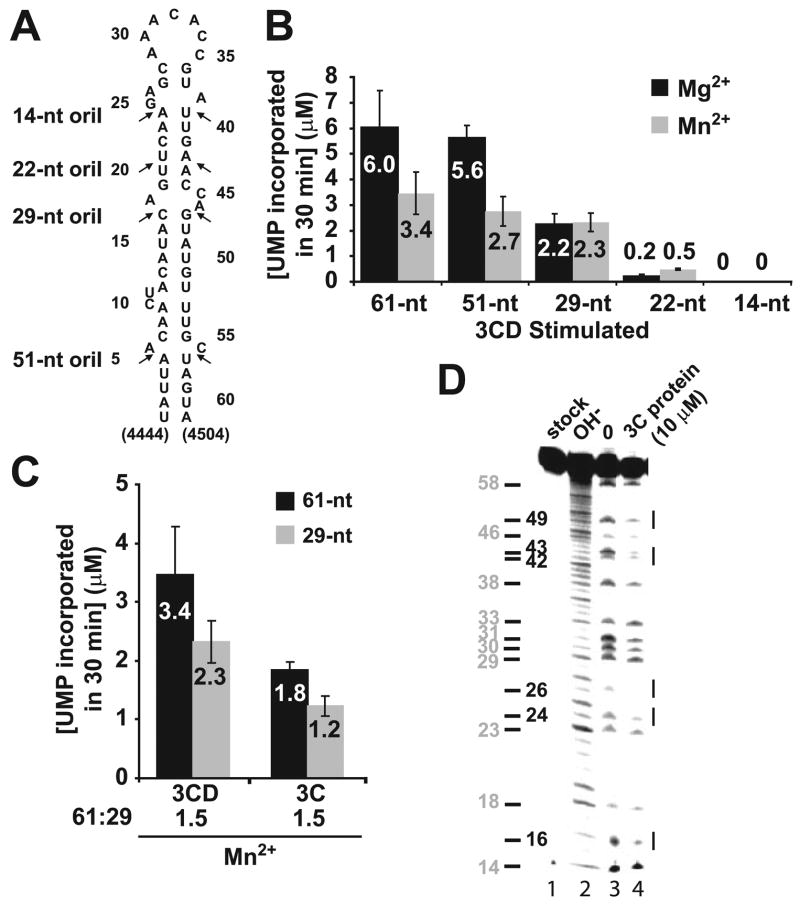
FIGURE 2. A 29-nt oriI element and protein 3C are sufficient to direct specific and efficient VPg uridylylation in vitro.
A, sites of oriI truncation. PV oriIs containing stems of various lengths as indicated were produced by using in vitro transcription (61 and 51 nt) or chemical synthesis (29, 22, and 14 nt). B, a 29-nt oriI is sufficient for VPg uridylylation in vitro. Shown is product formed in reactions containing oriI templates of varying length (1 μM) in the presence of Mg2+ (black bars) or Mn2+ (gray bars) as the cofactor for PV 3Dpol. Reactions were stimulated using 3CD (1 μM) in the presence of 3D (1 μM) and the indicated RNAs (1 μM). The average of at least two independent experiments is reported along with the corresponding S.D. values. C, 3C is sufficient for VPg uridylylation in vitro. Shown is product formed in reactions containing the 61-nt (black bars) or 29-nt oriI (gray bars) stimulated by using either 3CD or 3C. The average of at least two independent experiments is reported along with the corresponding S.D. values. D, footprint of 3C on oriI. Shown is iodine cleavage of AMPαS-substituted 61-nt oriI. Cleavage products were analyzed by denaturing PAGE. Shown is the phosphor image of the gel. Lane 1, untreated oriI; lane 2, oriI cleaved by alkaline hydrolysis; lane 3, iodine-treated oriI in the absence of 3C; lane 4, iodine-treated RNA in the presence of 3C (10 μM). Sites of 3C binding are indicated by lines to the right of the gel. The position of the adenosine residues is indicated to the left of the gel.